9KSJ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 8 | | Descriptor: | 3-[[(6E)-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-[5-(2-methoxyethoxy)pyridin-3-yl]-2,4-bis(oxidanylidene)-1,3,5-triazinan-1-yl]methyl]-4-methyl-benzenecarbonitrile, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
6RNW
 
 | |
6R46
 
 | |
6R48
 
 | |
6RNV
 
 | |
2NS2
 
 | | Crystal Structure of Spindlin1 | | Descriptor: | PHOSPHATE ION, Spindlin-1 | | Authors: | Zhao, Q, Qin, L, Jiang, F, Wu, B, Yue, W, Xu, F, Rong, Z, Yuan, H, Xie, X, Gao, Y, Bai, C, Bartlam, M. | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human spindlin1. Tandem tudor-like domains for cell cycle regulation
J.Biol.Chem., 282, 2007
|
|
8WCW
 
 | |
8WCX
 
 | |
6KSB
 
 | |
6LQ3
 
 | |
6LQ7
 
 | |
6LQ8
 
 | |
6LPY
 
 | |
6LQ5
 
 | |
5F5M
 
 | | Crystal structure of Marburg virus nucleoprotein core domain | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
5F5O
 
 | | Crystal structure of Marburg virus nucleoprotein core domain bound to VP35 regulation peptide | | Descriptor: | Nucleoprotein, Peptide from Polymerase cofactor VP35, SULFATE ION | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
4G3Y
 
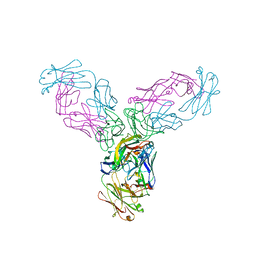 | | Crystal structure of TNF-alpha in complex with Infliximab Fab fragment | | Descriptor: | Tumor necrosis factor, infliximab Fab H, infliximab Fab L | | Authors: | Liang, S.Y, Dai, J.X, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2012-07-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for treating tumor necrosis factor alpha (TNFalpha)-associated diseases with the therapeutic antibody infliximab
J.Biol.Chem., 288, 2013
|
|
7XYD
 
 | | Crystal structure of TMPRSS2 in complex with Nafamostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
7Y0F
 
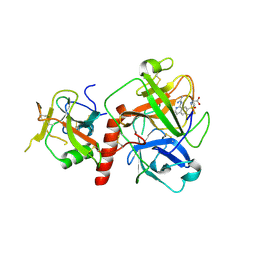 | | Crystal structure of TMPRSS2 in complex with UK-371804 | | Descriptor: | 2-[(1-carbamimidamido-4-chloranyl-isoquinolin-7-yl)sulfonylamino]-2-methyl-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, H, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-04 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
7Y0E
 
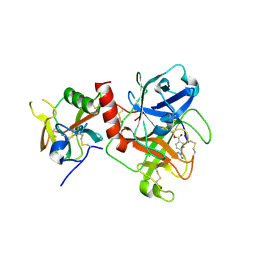 | | Crystal structure of TMPRSS2 in complex with Camostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-04 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
9IQE
 
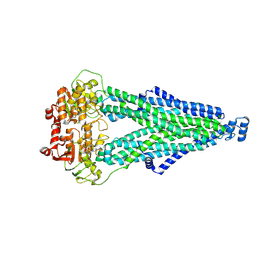 | |
9IQF
 
 | |
9IQG
 
 | | Cryo-EM structure of MsRv1273c/72c from Mycobacterium smegmatis in the ATP|ADP+Vi-bound Occ (Vi) state | | Descriptor: | ABC transporter transmembrane region, ABC transporter, ATP-binding protein, ... | | Authors: | Lan, Y, Yu, J, Li, J. | | Deposit date: | 2024-07-12 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of a mycobacterial isoniazid efflux pump MsRv1273c/72c with a degenerate nucleotide-binding site.
Nat Commun, 16, 2025
|
|
9JAL
 
 | | Cryo-EM structure of MPXV core protease in complex with compound A1 | | Descriptor: | A1ECM-DI8-ALA-ETA, Core protease I7 | | Authors: | Gao, Y, Xie, X, Lan, W, Wang, W, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
9JAN
 
 | | Cryo-EM structure of MPXV protease in complex with compound A4 | | Descriptor: | A1ECL-DI8-ALA-AEM, Core protease I7 | | Authors: | Gao, Y, Xie, X, Lan, W, Wang, W, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
