1D12
 
 | | STRUCTURAL COMPARISON OF ANTICANCER DRUG-DNA COMPLEXES. ADRIAMYCIN AND DAUNOMYCIN | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), DOXORUBICIN, SODIUM ION, ... | | Authors: | Frederick, C.A, Williams, L.D, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin.
Biochemistry, 29, 1990
|
|
1D13
 
 | | MOLECULAR STRUCTURE OF AN A-DNA DECAMER D(ACCGGCCGGT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Frederick, C.A, Quigley, G.J, Teng, M.-K, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of an A-DNA decamer d(ACCGGCCGGT).
Eur.J.Biochem., 181, 1989
|
|
1D14
 
 | | STRUCTURE OF 11-DEOXYDAUNOMYCIN BOUND TO DNA CONTAINING A PHOSPHOROTHIOATE | | Descriptor: | 6-DEOXYDAUNOMYCIN, DNA (5'-D(*CP*GP*TP*(AS)P*CP*G)-3') | | Authors: | Williams, L.D, Egli, M, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Quigley, G.J, Wang, A.H.-J, Rich, A, Frederick, C.A. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of 11-deoxydaunomycin bound to DNA containing a phosphorothioate.
J.Mol.Biol., 215, 1990
|
|
1D10
 
 | | STRUCTURAL COMPARISON OF ANTICANCER DRUG-DNA COMPLEXES. ADRIAMYCIN AND DAUNOMYCIN | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), SODIUM ION, ... | | Authors: | Frederick, C.A, Williams, L.D, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin.
Biochemistry, 29, 1990
|
|
1DNE
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 2 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-09-14 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment.
Biochemistry, 28, 1989
|
|
1AC7
 
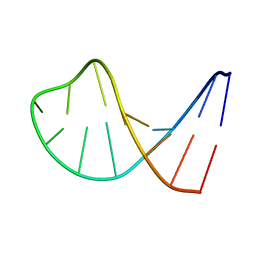 | | STRUCTURAL FEATURES OF THE DNA HAIRPIN D(ATCCTAGTTATAGGAT): THE FORMATION OF A G-A BASE PAIR IN THE LOOP, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*TP*AP*GP*TP*TP*AP*TP*AP*GP*GP*AP*T)-3') | | Authors: | Van Dongen, M.J.P, Mooren, M.M.W, Willems, E.F.A, Van Der Marel, G.A, Van Boom, J.H, Wijmenga, S.S, Hilbers, C.W. | | Deposit date: | 1997-02-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural features of the DNA hairpin d(ATCCTA-GTTA-TAGGAT): formation of a G-A base pair in the loop.
Nucleic Acids Res., 25, 1997
|
|
1D96
 
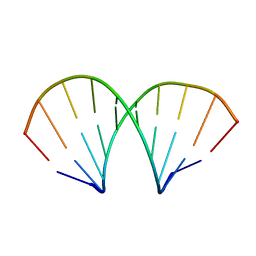 | | MOLECULAR STRUCTURE OF R(GCG)D(TATACGC): A DNA-RNA HYBRID HELIX JOINED TO DOUBLE HELICAL DNA | | Descriptor: | DNA/RNA (5'-R(*GP*CP*GP*)-D(*TP*AP*TP*AP*CP*GP*C)-3') | | Authors: | Wang, A.H.-J, Fujii, S, Van Boom, J.H, Van Der Marel, G.A, Van Boeckel, S.A.A, Rich, A. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of r(GCG)d(TATACGC): a DNA--RNA hybrid helix joined to double helical DNA.
Nature, 299, 1982
|
|
1B4Y
 
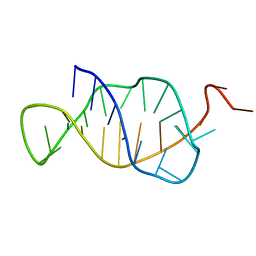 | | STRUCTURE AND MECHANISM OF FORMATION OF THE H-Y5 ISOMER OF AN INTRAMOLECULAR DNA TRIPLE HELIX. | | Descriptor: | DNA (H-Y5 TRIPLE HELIX) | | Authors: | Van Dongen, M.J.P, Doreleijers, J.F, Van Der Marel, G.A, Van Boom, J.H, Hilbers, C.W, Wijmenga, S.S. | | Deposit date: | 1998-12-30 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of formation of the H-y5 isomer of an intramolecular DNA triple helix.
Nat.Struct.Biol., 6, 1999
|
|
1D85
 
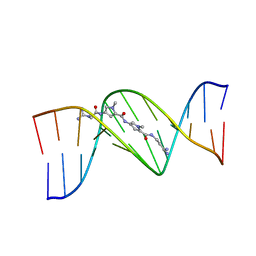 | | STRUCTURAL CONSEQUENCES OF A CARCINOGENIC ALKYLATION LESION ON DNA: EFFECT OF O6-ETHYL-GUANINE ON THE MOLECULAR STRUCTURE OF D(CGC[E6G]AATTCGCG)-NETROPSIN COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*(G36)P*AP*AP*TP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1992-08-24 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural consequences of a carcinogenic alkylation lesion on DNA: effect of O6-ethylguanine on the molecular structure of the d(CGC[e6G]AATTCGCG)-netropsin complex.
Biochemistry, 31, 1992
|
|
1D35
 
 | | FACILE FORMATION OF A CROSSLINKED ADDUCT BETWEEN DNA AND THE DAUNORUBICIN DERIVATIVE MAR70 MEDIATED BY FORMALDEHYDE: MOLECULAR STRUCTURE OF THE MAR70-D(CGTNACG) COVALENT ADDUC | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*(A40)P*CP*G)-3'), MAGNESIUM ION | | Authors: | Gao, Y.-G, Liaw, Y.-C, Li, Y.-K, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1D36
 
 | | FACILE FORMATION OF A CROSSLINKED ADDUCT BETWEEN DNA AND THE DAUNORUBICIN DERIVATIVE MAR70 MEDIATED BY FORMALDEHYDE: MOLECULAR STRUCTURE OF THE MAR70-D(CGTNACG) COVALENT ADDUC | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MAGNESIUM ION | | Authors: | Gao, Y.-G, Liaw, Y.-C, Li, Y.-K, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1D86
 
 | | STRUCTURAL CONSEQUENCES OF A CARCINOGENIC ALKYLATION LESION ON DNA: EFFECT OF O6-ETHYL-GUANINE ON THE MOLECULAR STRUCTURE OF D(CGC[E6G]AATTCGCG)-NETROPSIN COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1992-08-24 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural consequences of a carcinogenic alkylation lesion on DNA: effect of O6-ethylguanine on the molecular structure of the d(CGC[e6G]AATTCGCG)-netropsin complex.
Biochemistry, 31, 1992
|
|
7AKR
 
 | |
7AQM
 
 | | ADP-ribosylserine hydrolase ARH3 of Latimeria chalumnae in complex with alpha-1''-O-methyl-ADP-ribose (meADPr) | | Descriptor: | ADP-ribosylhydrolase like 2, Adenosine 5'-diphosphoric acid beta-[(3beta,4beta-dihydroxy-5beta-methoxytetrahydrofuran-2alpha-yl)methyl] estere, MAGNESIUM ION | | Authors: | Rack, J.G.M, Zorzini, V, Ahel, I. | | Deposit date: | 2020-10-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Nat Commun, 12, 2021
|
|
7AKS
 
 | |
7ARW
 
 | | Structure of human ARH3 E41A bound to alpha-NAD+ and magnesium | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADP-ribose glycohydrolase ARH3, ... | | Authors: | Rack, J.G.M, Zorzini, V, Ahel, I. | | Deposit date: | 2020-10-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Nat Commun, 12, 2021
|
|
7BZL
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-04-28 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8PEJ
 
 | | CjGH35 with a Galactosidase Activity-Based Probe | | Descriptor: | (1~{S},2~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W.A, Davies, G.J. | | Deposit date: | 2023-06-14 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The development of a broad-spectrum retaining beta-exo-galactosidase activity-based probe.
Org.Biomol.Chem., 21, 2023
|
|
4WSJ
 
 | | Crystal structure of a bacterial fucodiase in complex with 1-((1R,2R,3R,4R,5R,6R)-2,3,4-trihydroxy-5-methyl-7-azabicyclo[4.1.0]heptan-7-yl)ethan-1-one | | Descriptor: | Alpha-L-fucosidase, N-[(1S,2R,3R,4S,5R)-3,4,5-trihydroxy-2-methylcyclohexyl]acetamide, SULFATE ION | | Authors: | Davies, G.J, Wright, D.W. | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | In vitroandin vivocomparative and competitive activity-based protein profiling of GH29 alpha-l-fucosidases.
Chem Sci, 6, 2015
|
|
4WSK
 
 | | Crystal structure of a bacterial fucosidase with phenyl((1R,2R,3R,4R,5R,6R)-2,3,4-trihydroxy-5-methyl-7-azabicyclo[4.1.0]heptan-7-yl)methanone | | Descriptor: | Alpha-L-fucosidase, IMIDAZOLE, N-[(1S,2R,3R,4S,5R)-3,4,5-trihydroxy-2-methylcyclohexyl]benzamide, ... | | Authors: | Davies, G.J. | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | In vitroandin vivocomparative and competitive activity-based protein profiling of GH29 alpha-l-fucosidases.
Chem Sci, 6, 2015
|
|
7P2Z
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with cyclosulfamidate 4 | | Descriptor: | (3~{a}~{R},4~{S},5~{S},6~{S},7~{R},7~{a}~{S})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydro-3~{H}-benzo[d][1,2,3]oxathiazole-4,5,6-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Kok, K, Overkleeft, H, Artola, M, Sulzenbacher, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
7P32
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with cyclosulfamidate 6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Kok, K, Overkleeft, H, Artola, M, Sulzenbacher, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
7P4C
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with noncovalent Cyclophellitol Sulfamidate probe KK131 | | Descriptor: | (3aR,4S,5S,6R,7R,7aS)-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3a,4,5,6,7,7a-hexahydro-3H-benzo[d][1,2,3]oxathiazole-4,5,6-triol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
7P4D
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with covalent Cyclophellitol Sulfamidate probe KK130 | | Descriptor: | 1,2-ETHANEDIOL, OXALATE ION, Oligosaccharide 4-alpha-D-glucosyltransferase, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
8RW3
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase, complexed with a non-covalent 1,2- Cyclophellitol aziridine | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Moran, E, Davies, G, Ofamn, T, Heming, J, Nin-Hill, A, Kullmer, F, Steneker, R, Klein, A, Bennett, M, Ruijgrok, G, Kok, K, Aerts, J, Van der Marel, G, Rovira, C, Artola, M, Codee, J, Overkleeft, H. | | Deposit date: | 2024-02-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational and Electronic Variations in 1,2- and 1,5a-Cyclophellitols and their Impact on Retaining alpha-Glucosidase Inhibition.
Chemistry, 30, 2024
|
|
