7D6V
 
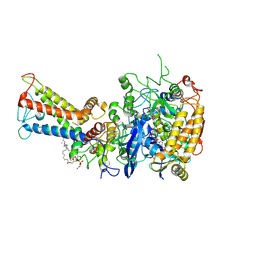 | | Mycobacterium smegmatis Sdh1 in complex with UQ1 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Zhou, X, Gao, Y, Wang, Q, Gong, H, Rao, Z. | | Deposit date: | 2020-10-02 | | Release date: | 2021-04-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Architecture of the mycobacterial succinate dehydrogenase with a membrane-embedded Rieske FeS cluster.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
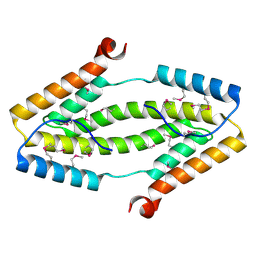
3ITF
 
 | |
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
6J6M
 
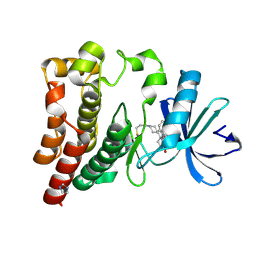 | | Co-crystal structure of BTK kinase domain with Zanubrutinib | | Descriptor: | (7S)-2-(4-phenoxyphenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, IMIDAZOLE, Tyrosine-protein kinase BTK | | Authors: | Zhou, X, Hong, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Zanubrutinib (BGB-3111), a Novel, Potent, and Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 62, 2019
|
|
5WWL
 
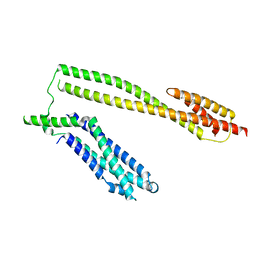 | | Crystal structure of the Schizogenesis pombe kinetochore Mis12C subcomplex | | Descriptor: | Centromere protein mis12, Kinetochore protein nnf1 | | Authors: | Wang, C, Zhou, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2017-01-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphorylation of CENP-C by Aurora B facilitates kinetochore attachment error correction in mitosis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8ESR
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 2 | | Descriptor: | 25S rRNA (cytosine-C(5))-methyltransferase nop2, 60S ribosomal protein L13, 60S ribosomal protein L14, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUG
 
 | | Ytm1 associated nascent 60S ribosome State 3 | | Descriptor: | 60S ribosomal protein L10-A, 60S ribosomal protein L11-A, 60S ribosomal protein L13, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUP
 
 | | Ytm1 associated 60S nascent ribosome State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ESQ
 
 | | Ytm1 associated nascent 60S ribosome State 2 | | Descriptor: | 25S rRNA (cytosine-C(5))-methyltransferase nop2, 60S ribosomal protein L13, 60S ribosomal protein L14, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EV3
 
 | | Ytm1 associated 60S nascent ribosome (-Fkbp39) State 1B | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETJ
 
 | | Fkbp39 associated 60S nascent ribosome State 2 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETC
 
 | | Fkbp39 associated nascent 60S ribosome State 4 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUY
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETI
 
 | | Fkbp39 associated 60S nascent ribosome State 1 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETG
 
 | | Fkbp39 associated 60S nascent ribosome State 3 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUI
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 3 | | Descriptor: | 60S ribosomal protein L10-A, 60S ribosomal protein L11-A, 60S ribosomal protein L13, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETH
 
 | | Ytm1 associated 60S nascent ribosome State 1B | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
7DPT
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, ADENOSINE-5'-DIPHOSPHATE, CTP synthase, ... | | Authors: | Liu, J.L, Zhou, X, Guo, C.J, Chang, C.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DPW
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Liu, J.L, Zhou, X, Guo, C.J, Chang, C.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4UIS
 
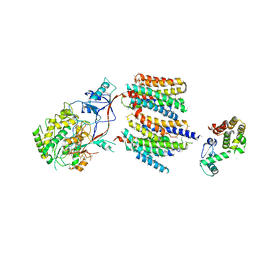 | | The cryoEM structure of human gamma-Secretase complex | | Descriptor: | GAMMA-SECRETASE, LYSOZYME | | Authors: | Sun, L, Zhao, L, Yang, G, Yan, C, Zhou, R, Zhou, X, Xie, T, Zhao, Y, Wu, S, Li, X, Shi, Y. | | Deposit date: | 2015-04-03 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis of Human Gamma-Secretase Assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4LA1
 
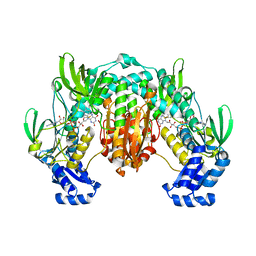 | | Crystal structure of SjTGR (thioredoxin glutathione reductase from Schistosoma japonicumi)complex with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase | | Authors: | Peng, Y, Wu, Q, Huang, F, Chen, J, Li, X, Zhou, X, Fan, X. | | Deposit date: | 2013-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structure of SjTGR complex with FAD
To be Published
|
|
6LUM
 
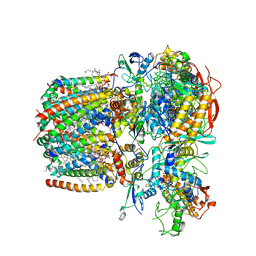 | | Structure of Mycobacterium smegmatis succinate dehydrogenase 2 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Gao, Y, Gong, H, Zhou, X, Xiao, Y, Wang, W, Ji, W, Wang, Q, Rao, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF.
Nat Commun, 11, 2020
|
|
5U6I
 
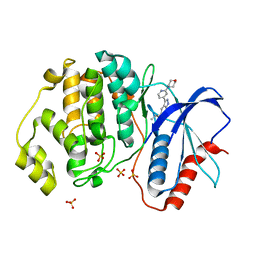 | | Discovery of MLi-2, an Orally Available and Selective LRRK2 Inhibitor that Reduces Brain Kinase Activity | | Descriptor: | 3-[2-(morpholin-4-yl)pyridin-4-yl]-5-[(propan-2-yl)oxy]-1H-indazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Scott, J.D, DeMong, D.E, Fell, M.J, Mirescu, C, Basu, K, Greshock, T.J, Morrow, J.A, Xiao, L, Hruza, A, Harris, J, Tiscia, H.E, Chang, R.K, Embrey, M.W, McCauley, J.A, Li, W, Lin, S, Liu, H, Dai, X, Baptista, M, Agnihotri, G, Columbus, J, Mei, H, Poirier, M, Zhou, X, Lin, Y, Yin, Z, Sanders, J.M, Drolet, R.E, Kern, J.T, Kennedy, M.E, Parker, E.M, Stamford, A.W, Nargund, R, Miller, M.W. | | Deposit date: | 2016-12-08 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a 3-(4-Pyrimidinyl) Indazole (MLi-2), an Orally Available and Selective Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitor that Reduces Brain Kinase Activity.
J. Med. Chem., 60, 2017
|
|
6LGY
 
 | | Crystal structure of a cysteine-pair mutant (P10C-S291C) of a bacterial bile acid transporter in an inward-facing state complexed with glycine and sodium | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, GLYCINE, NONAETHYLENE GLYCOL, ... | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Substrate binding in the bile acid transporter ASBT Yf from Yersinia frederiksenii.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6LH0
 
 | | Crystal structure of a cysteine-pair mutant (P10C-S291C) of a bacterial bile acid transporter in an inward-facing apo-state | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Transporter, sodium/bile acid symporter family | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.812 Å) | | Cite: | Substrate binding in the bile acid transporter ASBT Yf from Yersinia frederiksenii.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
