6ZLX
 
 | | The structure of the ClpX-associated factor PDIP38 | | Descriptor: | PLATINUM (II) ION, Polymerase delta-interacting protein 2 | | Authors: | Zeth, K, Dougan, D. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.394 Å) | | Cite: | PDIP38 is a novel adaptor-like modulator of the mitochondrial AAA+ protease CLPXP
To Be Published
|
|
7PDC
 
 | |
6FHG
 
 | |
6Z4E
 
 | | The structure of the C-terminal domain of RssB from E. coli | | Descriptor: | Regulator of RpoS | | Authors: | Zeth, K, Dimce, M, Terrence, D.M, Schuenemann, V, Dougan, D. | | Deposit date: | 2020-05-25 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into the RssB-Mediated Recognition and Delivery of sigma s to the AAA+ Protease, ClpXP.
Biomolecules, 10, 2020
|
|
6Z4C
 
 | | The structure of the N-terminal domain of RssB from E. coli | | Descriptor: | Regulator of RpoS | | Authors: | Zeth, K, Dimce, M, Terrence, D.M, Schuenemann, V, Dougan, D. | | Deposit date: | 2020-05-25 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into the RssB-Mediated Recognition and Delivery of sigma s to the AAA+ Protease, ClpXP.
Biomolecules, 10, 2020
|
|
3H7Z
 
 | |
3H7X
 
 | |
3ZRX
 
 | |
3ZRW
 
 | | The structure of the dimeric Hamp-Dhp fusion A291V mutant | | Descriptor: | AF1503 PROTEIN, OSMOLARITY SENSOR PROTEIN ENVZ | | Authors: | Zeth, K, Hulko, M, Ferris, H.U, Martin, J, Lupas, A.N. | | Deposit date: | 2011-06-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Regulation of Receptor Histidine Kinases.
Structure, 20, 2012
|
|
3ZX6
 
 | | Structure of Hamp(AF1503)-Tsr fusion - Hamp (A291V) mutant | | Descriptor: | CHLORIDE ION, HAMP, METHYL-ACCEPTING CHEMOTAXIS PROTEIN I | | Authors: | Zeth, K, Ferris, H.U, Lupas, A.L. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Axial Helix Rotation as a Mechanism for Signal Regulation Inferred from the Crystallographic Analysis of the E. Coli Serine Chemoreceptor.
J.Struct.Biol., 186, 2014
|
|
3ZRV
 
 | | The high resolution structure of a dimeric Hamp-Dhp fusion displays asymmetry - A291F mutant | | Descriptor: | HAMP, OSMOLARITY SENSOR PROTEIN ENVZ | | Authors: | Zeth, K, Hulko, M, Ferris, H.U, Martin, J. | | Deposit date: | 2011-06-20 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of Regulation of Receptor Histidine Kinases.
Structure, 20, 2012
|
|
4N59
 
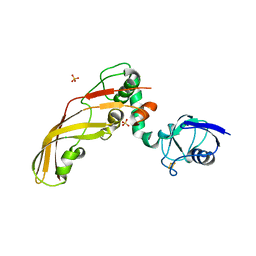 | | The Crystal Structure of Pectocin M2 at 2.3 Angstroms | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Pectocin M2, ... | | Authors: | Zeth, K, Grinter, R, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2013-10-09 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake.
Mol.Microbiol., 93, 2014
|
|
3LT6
 
 | |
3LT7
 
 | |
3D9X
 
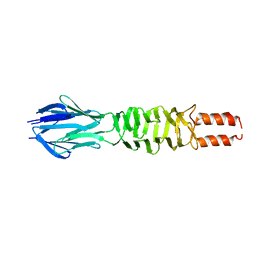 | |
5NNT
 
 | | The dimeric structure of LL-37 crystallized in DPC | | Descriptor: | Cathelicidin antimicrobial peptide, dodecyl 2-(trimethylammonio)ethyl phosphate | | Authors: | Zeth, K, Sancho-Vaello, E. | | Deposit date: | 2017-04-10 | | Release date: | 2018-01-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Structural remodeling and oligomerization of human cathelicidin on membranes suggest fibril-like structures as active species.
Sci Rep, 7, 2017
|
|
5NNK
 
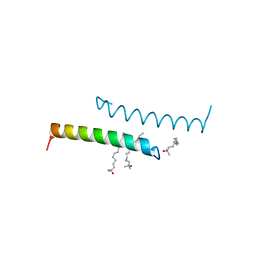 | | The structure of LL-37 crystallized in the presence LDAO | | Descriptor: | Cathelicidin antimicrobial peptide, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Zeth, K, Sancho-Vaello, E. | | Deposit date: | 2017-04-10 | | Release date: | 2018-01-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural remodeling and oligomerization of human cathelicidin on membranes suggest fibril-like structures as active species.
Sci Rep, 7, 2017
|
|
5NMN
 
 | |
5NNM
 
 | | The crystal structure of dimeric LL-37 | | Descriptor: | CARBONATE ION, Cathelicidin antimicrobial peptide | | Authors: | Zeth, K, Sancho-Vaello, E. | | Deposit date: | 2017-04-10 | | Release date: | 2018-01-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural remodeling and oligomerization of human cathelicidin on membranes suggest fibril-like structures as active species.
Sci Rep, 7, 2017
|
|
2FGR
 
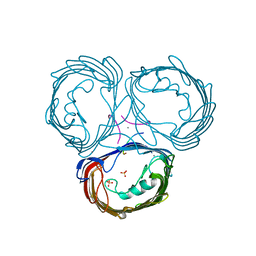 | | High resolution Xray structure of Omp32 | | Descriptor: | CALCIUM ION, Outer membrane porin protein 32, PAP, ... | | Authors: | Zeth, K, Zachariae, U, Engelhardt, H. | | Deposit date: | 2005-12-22 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution crystal structures and molecular dynamics studies reveal substrate binding in the porin omp32
J.Biol.Chem., 281, 2006
|
|
1TE0
 
 | |
4C4V
 
 | |
4ARM
 
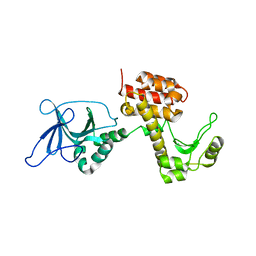 | | Structure of the inactive pesticin T201A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARL
 
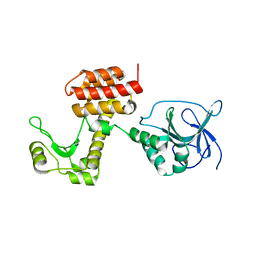 | | Structure of the inactive pesticin D207A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4ARQ
 
 | | Structure of the pesticin S89C, S285C double mutant | | Descriptor: | MAGNESIUM ION, PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
