7L61
 
 | |
7L67
 
 | |
3OUW
 
 | | Structure of beta-catenin with Lef-1 | | Descriptor: | Catenin beta-1, Lymphoid enhancer-binding factor 1 | | Authors: | Weis, W.I, Sun, J. | | Deposit date: | 2010-09-15 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Biochemical and structural characterization of beta-catenin interactions with nonphosphorylated and CK2-phosphorylated Lef-1.
J.Mol.Biol., 405, 2011
|
|
3OUX
 
 | | Structure of beta-catenin with phosphorylated Lef-1 | | Descriptor: | Catenin beta-1, Lymphoid enhancer-binding factor 1 | | Authors: | Weis, W.I, Sun, J. | | Deposit date: | 2010-09-15 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural characterization of beta-catenin interactions with nonphosphorylated and CK2-phosphorylated Lef-1.
J.Mol.Biol., 405, 2011
|
|
7JUH
 
 | |
7JUG
 
 | |
7JUB
 
 | |
7JUF
 
 | |
7JUE
 
 | |
7JUD
 
 | |
7JUC
 
 | |
4U16
 
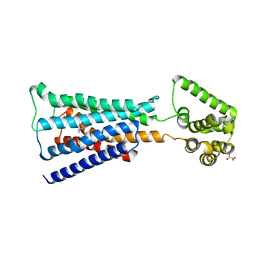 | | M3-mT4L receptor bound to NMS | | Descriptor: | D(-)-TARTARIC ACID, Muscarinic acetylcholine receptor M3,Lysozyme,Muscarinic acetylcholine receptor M3, N-methyl scopolamine | | Authors: | Thorsen, T.S, Matt, R, Weis, W.I, Kobilka, B. | | Deposit date: | 2014-07-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
3CF2
 
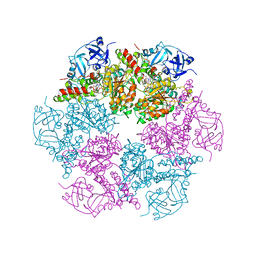 | | Structure of P97/vcp in complex with ADP/AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Davies, J.M, Delabarre, B, Brunger, A.T, Weis, W.I. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change.
Structure, 16, 2008
|
|
6WVT
 
 | | Structural basis of alphaE-catenin - F-actin catch bond behavior | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Xu, X.P, Pokutta, S, Torres, M, Swift, M.F, Hanein, D, Volkmann, N, Weis, W.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-10-07 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of alpha E-catenin-F-actin catch bond behavior.
Elife, 9, 2020
|
|
1KMB
 
 | | SELECTIN-LIKE MUTANT OF MANNOSE-BINDING PROTEIN A | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-A | | Authors: | Ng, K.K.-S, Weis, W.I. | | Deposit date: | 1996-11-07 | | Release date: | 1997-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a selectin-like mutant of mannose-binding protein complexed with sialylated and sulfated Lewis(x) oligosaccharides.
Biochemistry, 36, 1997
|
|
3KMB
 
 | |
1YTT
 
 | | YB SUBSTITUTED SUBTILISIN FRAGMENT OF MANNOSE BINDING PROTEIN-A (SUB-MBP-A), MAD STRUCTURE AT 110K | | Descriptor: | MANNOSE-BINDING PROTEIN A, YTTERBIUM (III) ION | | Authors: | Burling, F.T, Weis, W.I, Flaherty, K.M, Brunger, A.T. | | Deposit date: | 1995-11-09 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of protein solvation and discrete disorder with experimental crystallographic phases.
Science, 271, 1996
|
|
9QY1
 
 | | Endo180 (uPARAP) carbohydrate-recognition domain 2 with bound methyl fucoside | | Descriptor: | CALCIUM ION, Mannose receptor C type 2, methyl alpha-L-fucopyranoside | | Authors: | Drickamer, K, Feinberg, H, Weis, W.I, Taylor, M.E, Jegouzo, S.A.F. | | Deposit date: | 2025-04-16 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sugar binding activity of the endothelial receptor Endo180
To Be Published
|
|
1T08
 
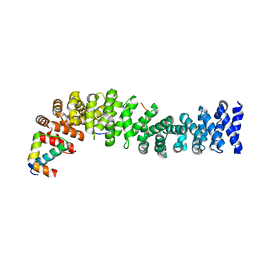 | | Crystal structure of beta-catenin/ICAT helical domain/unphosphorylated APC R3 | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin, Beta-catenin-interacting protein 1 | | Authors: | Ha, N.-C, Tonozuka, T, Stamos, J.L, Weis, W.I. | | Deposit date: | 2004-04-07 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation
Mol.Cell, 15, 2004
|
|
2RH1
 
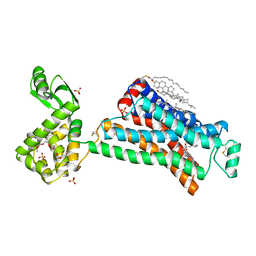 | | High resolution crystal structure of human B2-adrenergic G protein-coupled receptor. | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Cherezov, V, Rosenbaum, D.M, Hanson, M.A, Rasmussen, S.G.F, Thian, F.S, Kobilka, T.S, Choi, H.J, Kuhn, P, Weis, W.I, Kobilka, B.K, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2007-10-05 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor.
Science, 318, 2007
|
|
9EJK
 
 | |
9EJL
 
 | |
9EJM
 
 | | Lgl2 bound to the aPKCiota-Par6B complex in its ADP-bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, LLGL scribble cell polarity complex component 2, MAGNESIUM ION, ... | | Authors: | Almagor, L, Weis, W.I. | | Deposit date: | 2024-11-28 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Polarity protein Par6 facilitates the processive phosphorylation of Lgl via a dynamic interaction with aPKC.
Commun Biol, 8, 2025
|
|
3C98
 
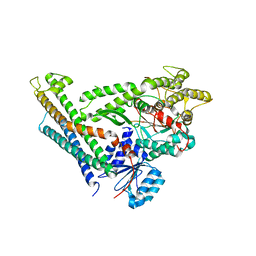 | | Revised structure of the munc18a-syntaxin1 complex | | Descriptor: | Syntaxin-1A, Syntaxin-binding protein 1 | | Authors: | Hattendorf, D.A, Misura, K.M, Burkhardt, P, Scheller, R.H, Fasshauer, D, Weis, W.I. | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide
Embo J., 27, 2008
|
|
4KRR
 
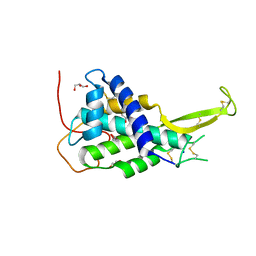 | | Crystal structure of Drosophila WntD N-terminal domain-linker (residues 31-240) | | Descriptor: | GLYCEROL, SODIUM ION, Wnt inhibitor of Dorsal protein | | Authors: | Chu, M.L.-H, Choi, H.-J, Ahn, V.E, Daniels, D.L, Nusse, R, Weis, W.I. | | Deposit date: | 2013-05-16 | | Release date: | 2013-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Structural Studies of Wnts and Identification of an LRP6 Binding Site.
Structure, 21, 2013
|
|
