4AGF
 
 | |
4AGE
 
 | |
5AGC
 
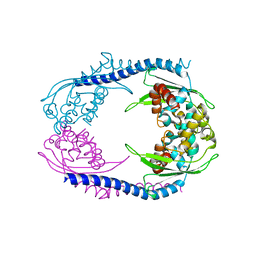 | | Crystallographic forms of the Vps75 tetramer | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 75 | | Authors: | Hammond, C.M, Sundaramoorthy, R, Owen-Hughes, T. | | Deposit date: | 2015-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Histone Chaperone Vps75 Forms Multiple Oligomeric Assemblies Capable of Mediating Exchange between Histone H3-H4 Tetramers and Asf1-H3-H4 Complexes.
Nucleic Acids Res., 44, 2016
|
|
3LCC
 
 | |
6TO3
 
 | |
8ATG
 
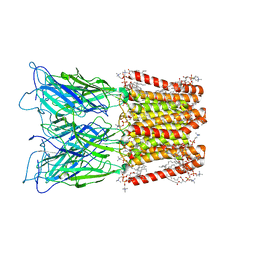 | | Pentameric ligand-gated ion channel GLIC with bound lipids | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Proton-gated ion channel | | Authors: | Bergh, C, Rovsnik, U, Howard, R.J, Lindahl, E. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of lipid binding sites in a ligand-gated ion channel by integrating simulations and cryo-EM.
Elife, 12, 2024
|
|
5EL8
 
 | |
8SBE
 
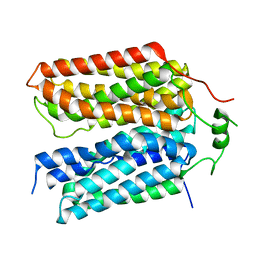 | | Structure of the rat vesicular glutamate transporter 2 determined by single-particle Cryo-EM | | Descriptor: | Vesicular glutamate transporter 2 | | Authors: | Li, F, Finer-Moore, J, Eriksen, J, Cheng, Y, Edwards, R, Stroud, R. | | Deposit date: | 2023-04-03 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ion transport and regulation in a synaptic vesicle glutamate transporter.
Science, 368, 2020
|
|
7OBO
 
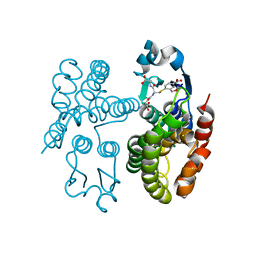 | | GSTF1 from Alopecurus myosuroides | | Descriptor: | (2~{S})-2-azanyl-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-3-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]-1-oxidanylidene-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, Glutathione transferase | | Authors: | Pohl, E, Eno, R.F.M, Freitag-Pohl, S. | | Deposit date: | 2021-04-23 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Flavonoid-based inhibitors of the Phi-class glutathione transferase from black-grass to combat multiple herbicide resistance.
Org.Biomol.Chem., 19, 2021
|
|
7ODM
 
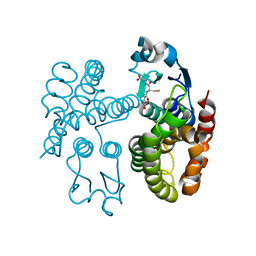 | | AmGSTF1 Y118S variant | | Descriptor: | Glutathione transferase, [(2~{S})-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-3-sulfanyl-propan-2-yl]amino]-1-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]azanium | | Authors: | Pohl, E, Eno, R.F.M. | | Deposit date: | 2021-04-30 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Flavonoid-based inhibitors of the Phi-class glutathione transferase from black-grass to combat multiple herbicide resistance.
Org.Biomol.Chem., 19, 2021
|
|
1E6B
 
 | |
8SXJ
 
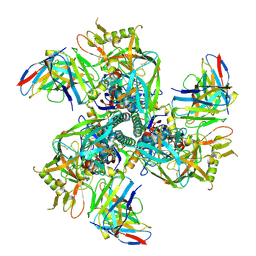 | | CH505 Disulfide Stapled SOSIP Bound to CH235.12 Fab | | Descriptor: | CH235.12 Heavy Chain, CH235.12 Light Chain, Envelope glycoprotein gp160, ... | | Authors: | Henderson, R. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv, 10, 2024
|
|
8SXI
 
 | | CH505 Disulfide Stapled SOSIP Bound to b12 Fab | | Descriptor: | Envelope glycoprotein gp160, HIV-1 gp41, b12 Heavy Chain, ... | | Authors: | Henderson, R. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv, 10, 2024
|
|
3L6D
 
 | | Crystal structure of putative oxidoreductase from Pseudomonas putida KT2440 | | Descriptor: | Putative oxidoreductase | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative oxidoreductase from Pseudomonas putida KT2440
To be Published
|
|
5ELA
 
 | |
5ELG
 
 | |
5SYD
 
 | |
3UEO
 
 | |
3UEN
 
 | | Crystal structure of TopBP1 BRCT4/5 domains | | Descriptor: | DNA topoisomerase 2-binding protein 1, GLYCEROL, THIOCYANATE ION | | Authors: | Leung, C.C, Glover, J.N.M. | | Deposit date: | 2011-10-31 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into recognition of MDC1 by TopBP1 in DNA replication checkpoint control.
Structure, 21, 2013
|
|
2YEK
 
 | |
2YEL
 
 | | Crystal Structure of the First Bromodomain of Human Brd4 with the inhibitor GW841819X | | Descriptor: | 1,2-ETHANEDIOL, BENZYL [(4R)-1-METHYL-6-PHENYL-4H-[1,2,4]TRIAZOLO[4,3-A][1,4]BENZODIAZEPIN-4-YL]CARBAMATE, HUMAN BRD4 | | Authors: | Chung, C.W. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Characterization of Small Molecule Inhibitors of the Bet Family Bromodomains.
J.Med.Chem., 54, 2011
|
|
2YEM
 
 | |
3B2R
 
 | | Crystal Structure of PDE5A1 catalytic domain in complex with Vardenafil | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Huanchen, W, Mengchun, Y, Howard, R, Sharron, H.F, Hengming, K. | | Deposit date: | 2007-10-19 | | Release date: | 2008-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Conformational variations of both phosphodiesterase-5 and inhibitors provide the structural basis for the physiological effects of vardenafil and sildenafil.
Mol.Pharmacol., 73, 2008
|
|
3BJC
 
 | | Crystal structure of the PDE5A catalytic domain in complex with a novel inhibitor | | Descriptor: | 5-ethoxy-4-(1-methyl-7-oxo-3-propyl-6,7-dihydro-1H-pyrazolo[4,3-d]pyrimidin-5-yl)thiophene-2-sulfonamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, G, Wang, H, Howard, R, Cai, J, Wan, Y, Ke, H. | | Deposit date: | 2007-12-03 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An insight into the pharmacophores of phosphodiesterase-5 inhibitors from synthetic and crystal structural studies
BIOCHEM.PHARM., 75, 2008
|
|
5UA3
 
 | | Crystal Structure of a DNA G-Quadruplex with a Cytosine Bulge | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*GP*CP*GP*GP*AP*GP*GP*GP*TP*GP*GP*GP*CP*CP*T)-3'), POTASSIUM ION | | Authors: | Meier, M, McDougall, M.D, Krahn, N.J, Moya-Torres, A, McRae, E.K.S, Booy, E.P, Patel, T.R, McKenna, S.A, Stetefeld, J. | | Deposit date: | 2016-12-18 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8802 Å) | | Cite: | Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge.
Nucleic Acids Res., 46, 2018
|
|
