7KGQ
 
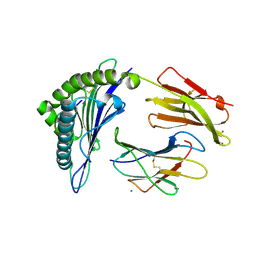 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N222-230 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGP
 
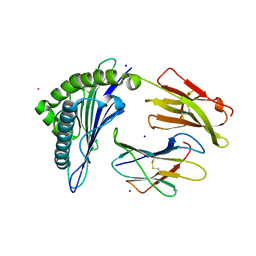 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N316-324 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGO
 
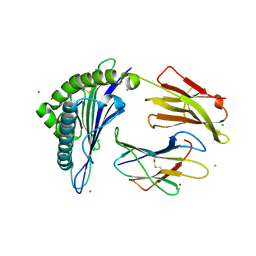 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N351-359 | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
1YIV
 
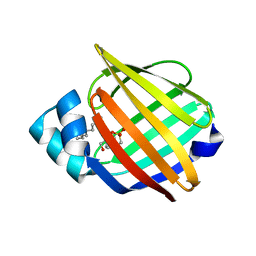 | | Structure of myelin P2 protein from Equine spinal cord | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LAURYL DIMETHYLAMINE-N-OXIDE, Myelin P2 protein | | Authors: | Hunter, D.J.B, MacMaster, R, Rozak, A.W, Riboldi-Tunnicliffe, A, Grifiths, I.R, Freer, A.A. | | Deposit date: | 2005-01-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of myelin P2 protein from equine spinal cord.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A9O
 
 | | Crystal structures of an activated YycF homologue, the essential response regulator from S.pneumoniae in complex with BeF3 and the effect of pH on BeF3 binding, possible phosphate in the active site | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MANGANESE (II) ION, Response regulator | | Authors: | Riboldi-Tunnicliffe, A, Isaacs, N.W, Mitchell, T.J. | | Deposit date: | 2005-07-12 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of an activated YycF homologue, the essential response regulator from S.pneumoniae in complex with BeF3 and the effect of pH on BeF3 binding, possible phosphate in the active site.
TO BE PUBLISHED
|
|
2A9R
 
 | | RR02-Rec Phosphate in the active site | | Descriptor: | DNA-binding response regulator, MAGNESIUM ION, XENON | | Authors: | Riboldi-Tunnicliffe, A. | | Deposit date: | 2005-07-12 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Crystal structures of an activated YycF homologue, the essential response regulator from S. pneumoniae in complex with BeF3 and the effect of pH on BeF3 binding, possible phosphate in the active site
To be Published
|
|
2A9P
 
 | | Medium Resolution BeF3 bound RR02-rec | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MANGANESE (II) ION, Response regulator | | Authors: | Riboldi-Tunnicliffe, A, Isaacs, N.W, Mitchell, T.J. | | Deposit date: | 2005-07-12 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of an activated YycF homologue, the essential response regulator from S.pneumoniae in complex with BeF3 and the effect of pH on BeF3 binding, possible phosphate in the active site.
TO BE PUBLISHED
|
|
2CS7
 
 | |
6I9F
 
 | | Solution structure of As-p18 reveals that nematode fatty acid binding proteins exhibit unusual structural features | | Descriptor: | Fatty acid-binding protein homolog, OLEIC ACID | | Authors: | Ibanez Shimabukuro, M, Rey Burusco, M.F, Kennedy, M.W, Corsico, B, Smith, B.O. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | As-p18, an extracellular fatty acid binding protein of nematodes, exhibits unusual structural features.
Biosci.Rep., 2019
|
|
8EB9
 
 | |
8EBB
 
 | |
7T69
 
 | | Crystal structure of Avr3 (SIX1) from Fusarium oxysporum f. sp. lycopersici | | Descriptor: | Avr3 (SIX1), Secreted in xylem 1, SULFATE ION | | Authors: | Yu, D.S, Outram, M.A, Ericsson, D.J, Jones, D.A, Williams, S.J. | | Deposit date: | 2021-12-13 | | Release date: | 2023-01-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structural repertoire of Fusarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies
Elife, 2023
|
|
7T6A
 
 | | Crystal structure of Avr1 (SIX4) from Fusarium oxysporum f. sp. lycopersici | | Descriptor: | Avr1 (FolSIX4), Avirulence protein 1, Avr1 (SIX4), ... | | Authors: | Yu, D.S, Outram, M.A, Ericsson, D.J, Jones, D.A, Williams, S.J. | | Deposit date: | 2021-12-13 | | Release date: | 2023-01-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structural repertoire of Fusarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies
Elife, 2023
|
|
8F2G
 
 | | Crystal structure of Hen Egg White Lysozyme at 0.44 GPa | | Descriptor: | Lysozyme C | | Authors: | Marshall, A.C, Boer, S.A, Turner, G, Moggach, S.A, Bond, C.S, Vrielink, A. | | Deposit date: | 2022-11-08 | | Release date: | 2022-12-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | High-pressure single-crystal diffraction at the Australian Synchrotron.
J.Synchrotron Radiat., 30, 2023
|
|
6W9Q
 
 | | Peptide-bound SARS-CoV-2 Nsp9 RNA-replicase | | Descriptor: | 3C-like proteinase peptide, Non-structural protein 9 fusion, PHOSPHATE ION | | Authors: | Littler, D.R, Gully, B.S, Riboldi-Tunnicliffe, A, Rossjohn, J. | | Deposit date: | 2020-03-23 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9.
Iscience, 23, 2020
|
|
6WC1
 
 | |
6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | Descriptor: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
6XAA
 
 | | SARS CoV-2 PLpro in complex with ubiquitin propargylamide | | Descriptor: | Non-structural protein 3, Ubiquitin-propargylamide, ZINC ION | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
7SUT
 
 | | Light harvesting phycobiliprotein HaPE645 from the cryptophyte Hemiselmis andersenii CCMP644 | | Descriptor: | (15,16)-DIHYDROBILIVERDIN (SINGLY LINKED), 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2021-11-18 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
7SSF
 
 | | Light harvesting phycobiliprotein HaPE560 from the cryptophyte Hemiselmis andersenii CCMP644 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DiCys-(15,16)-Dihydrobiliverdin, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2021-11-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii
Commun Biol, 2023
|
|
7LGD
 
 | | HLA-B*07:02 in complex with SARS-CoV-2 nucleocapsid peptide N105-113 | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Szeto, C, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | CD8 + T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses.
Immunity, 54, 2021
|
|
7LGT
 
 | | HLA-B*07:02 in complex with 229E-derived coronavirus nucleocapsid peptide N75-83 | | Descriptor: | BROMIDE ION, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Gras, S, Szeto, C, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-01-21 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | CD8 + T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses.
Immunity, 54, 2021
|
|
1NMT
 
 | |
4AK4
 
 | | High resolution structure of Galactose Binding lectin from Champedak (CGB) | | Descriptor: | AGGLUTININ ALPHA CHAIN, AGGLUTININ BETA-4 CHAIN, HEXAETHYLENE GLYCOL | | Authors: | Gabrielsen, M, Abdul-Rahman, P.S, Othman, S, Hashim, O.H, Cogdell, R.J. | | Deposit date: | 2012-02-21 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures and Binding Specificity of Galactose- and Mannose-Binding Lectins from Champedak: Differences from Jackfruit Lectins
Acta Crystallogr.,Sect.F, 70, 2014
|
|
7RTD
 
 | |
