3UAA
 
 | | Multicopper Oxidase CueO mutant C500SE506Q (data1) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An O-centered structure of the trinuclear copper center in the Cys500Ser/Glu506Gln mutant of CueO and structural changes in low to high X-ray dose conditions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3UAE
 
 | | Multicopper Oxidase CueO mutant C500SE506Q (data6) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An O-centered structure of the trinuclear copper center in the Cys500Ser/Glu506Gln mutant of CueO and structural changes in low to high X-ray dose conditions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3UAB
 
 | | Multicopper Oxidase CueO mutant C500SE506Q (data2) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An O-centered structure of the trinuclear copper center in the Cys500Ser/Glu506Gln mutant of CueO and structural changes in low to high X-ray dose conditions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3UAD
 
 | | Multicopper Oxidase CueO mutant C500SE506Q (data5) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An O-centered structure of the trinuclear copper center in the Cys500Ser/Glu506Gln mutant of CueO and structural changes in low to high X-ray dose conditions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1WG3
 
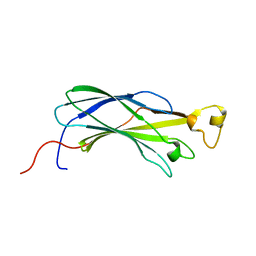 | | Structural analysis of yeast nucleosome-assembly factor CIA1p | | Descriptor: | Anti-silencing protein 1 | | Authors: | Padmanabhan, B, Kataoka, K, Umehara, T, Adachi, N, Yokoyama, S, Horikoshi, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Similarity between Histone Chaperone Cia1p/Asf1p and DNA-Binding Protein NF-{kappa}B
J.Biochem.(Tokyo), 138, 2005
|
|
8HEW
 
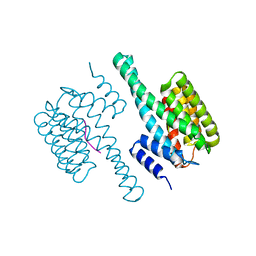 | | Potato 14-3-3 St14f | | Descriptor: | 14-3-3 protein, StFDL1 peptide | | Authors: | Taoka, K, Kawahara, I, Shinya, S, Harada, K, Muranaka, T, Furuita, K, Nakagawa, A, Fujiwara, T, Tsuji, H, Kojima, C. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Multifunctional chemical inhibitors of the florigen activation complex discovered by structure-based high-throughput screening.
Plant J., 112, 2022
|
|
7YOW
 
 | |
2YXV
 
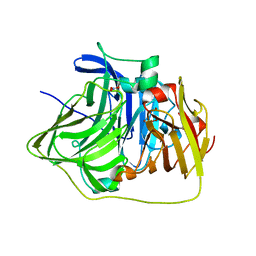 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
7WNB
 
 | |
7WN2
 
 | |
8I5I
 
 | |
8I5H
 
 | |
2YXW
 
 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
1RKR
 
 | | CRYSTAL STRUCTURE OF AZURIN-I FROM ALCALIGENES XYLOSOXIDANS NCIMB 11015 | | Descriptor: | AZURIN-I, COPPER (II) ION | | Authors: | Li, C, Inoue, T, Gotowda, M, Suzuki, S, Yamaguchi, K, Kataoka, K, Kai, Y. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of azurin I from the denitrifying bacterium Alcaligenes xylosoxidans NCIMB 11015 at 2.45 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6IQX
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - M467Q mutant, aerobically prepared | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Bilirubin oxidase, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
6IQZ
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - wild type | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
6IQY
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - M467Q mutant, anaerobically prepared | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Bilirubin oxidase, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
4E9Y
 
 | | Multicopper Oxidase mgLAC (data4) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Multicopper oxidase, ... | | Authors: | Komori, H, Miyazaki, K, Higuchi, Y. | | Deposit date: | 2012-03-21 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New insights into the catalytic active-site structure of multicopper oxidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4E9W
 
 | | Multicopper Oxidase mgLAC (data2) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Multicopper oxidase, ... | | Authors: | Komori, H, Miyazaki, K, Higuchi, Y. | | Deposit date: | 2012-03-21 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New insights into the catalytic active-site structure of multicopper oxidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4E9X
 
 | | Multicopper Oxidase mgLAC (data3) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Multicopper oxidase, ... | | Authors: | Komori, H, Miyazaki, K, Higuchi, Y. | | Deposit date: | 2012-03-21 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | New insights into the catalytic active-site structure of multicopper oxidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4E9V
 
 | | Multicopper Oxidase mgLAC (data1) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, HYDROXIDE ION, ... | | Authors: | Komori, H, Miyazaki, K, Higuchi, Y. | | Deposit date: | 2012-03-21 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New insights into the catalytic active-site structure of multicopper oxidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
1WS8
 
 | | Crystal Structure of Mavicyanin from Cucurbita pepo medullosa (Zucchini) | | Descriptor: | COPPER (II) ION, GLYCEROL, mavicyanin | | Authors: | Xie, Y, Inoue, T, Miyamoto, Y, Matsumura, H, Kunishige, K, Yamaguchi, K, Nojini, M, Suzuki, S, Kai, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural reorganization of the copper binding site involving Thr15 of mavicyanin from Cucurbita pepo medullosa (zucchini) upon reduction.
J.Biochem.(Tokyo), 137, 2005
|
|
2DV6
 
 | | Crystal structure of nitrite reductase from Hyphomicrobium denitrificans | | Descriptor: | COPPER (II) ION, Nitrite reductase, POTASSIUM ION | | Authors: | Nojiri, M, Xie, Y, Yamamoto, T, Inoue, T, Suzuki, S, Kai, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of a hexameric copper-containing nitrite reductase.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3VSO
 
 | | Human PPAR gamma ligand binding domain in complex with a gamma selective agonist mekt21 | | Descriptor: | (2R)-2-benzyl-3-[4-propoxy-3-({[4-(pyrimidin-2-yl)benzoyl]amino}methyl)phenyl]propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Waku, T, Ohashi, M, Morikawa, K, Miyachi, H. | | Deposit date: | 2012-04-30 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of a series of alpha-benzyl phenylpropanoic acid-type peroxisome proliferator-activated receptor (PPAR) gamma partial agonists with improved aqueous solubility
Bioorg.Med.Chem., 21, 2013
|
|
