2NLI
 
 | | Crystal Structure of the complex between L-lactate oxidase and a substrate analogue at 1.59 angstrom resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, HYDROGEN PEROXIDE, LACTIC ACID, ... | | Authors: | Furuichi, M, Suzuki, N, Balasundaresan, D, Yoshida, Y, Minagawa, H, Watanabe, Y, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2006-10-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
3F89
 
 | | NEMO CoZi domain | | Descriptor: | NF-kappa-B essential modulator | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
8KG3
 
 | | Structure of THOUSAND-GRAIN WEIGHT 6 (TGW6) | | Descriptor: | Os06g0623700 protein | | Authors: | Akabane, T, Suzuki, N, Matsumura, H, Yoshizawa, T, Tsuchiya, W, Katoh, E, Hirotsu, N. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | THOUSAND-GRAIN WEIGHT 6, which is an IAA-glucose hydrolase, preferentially recognizes the structure of the indole ring.
Sci Rep, 14, 2024
|
|
7BUX
 
 | | Eucommia ulmoides FPS1 | | Descriptor: | FPS2 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
7BUW
 
 | | Eucommia ulmoides TPT3 mutant -C94Y/A95F | | Descriptor: | FPS3 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
7BUV
 
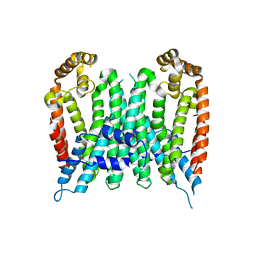 | | Eucommia ulmoides TPT3, crystal form 2 | | Descriptor: | FPS3 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
7BUU
 
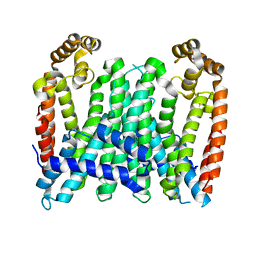 | | Eucommia ulmoides TPT3, crystal form 1 | | Descriptor: | FPS3 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
1WQZ
 
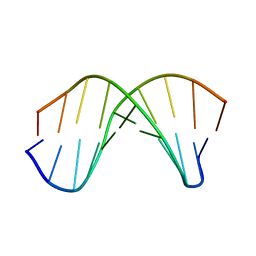 | | Complicated water orientations in the minor groove of B-DNA decamer D(CCATTAATGG)2 observed by neutron diffraction measurements | | Descriptor: | 5'-D(*CP*CP*AP*TP*TP*AP*AP*TP*GP*G)-3' | | Authors: | Arai, S, Chatake, T, Ohhara, T, Kurihara, K, Tanaka, I, Suzuki, N, Fujimoto, Z, Mizuno, H, Niimura, N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (3 Å) | | Cite: | Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements
Nucleic Acids Res., 33, 2005
|
|
1WQY
 
 | | X-RAY structural analysis of B-DNA decamer D(CCATTAATGG)2 crystal grown in D2O solution | | Descriptor: | 5'-D(*CP*CP*AP*TP*TP*AP*AP*TP*GP*G)-3' | | Authors: | Arai, S, Chatake, T, Ohhara, T, Kurihara, K, Tanaka, I, Suzuki, N, Fujimoto, Z, Mizuno, H, Niimura, N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements
Nucleic Acids Res., 33, 2005
|
|
1WLZ
 
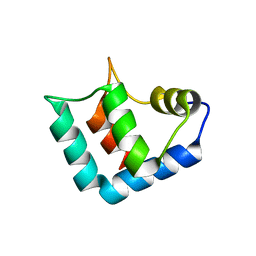 | | Crystal structure of DJBP fragment which was obtained by limited proteolysis | | Descriptor: | CAP-binding protein complex interacting protein 1 isoform a | | Authors: | Honbou, K, Suzuki, N, Horiuchi, M, Taira, T, Niki, T, Ariga, H, Inagaki, F. | | Deposit date: | 2004-07-01 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of DJBP Fragment which was obtained by Limited Proteolysis
To be Published
|
|
5X7H
 
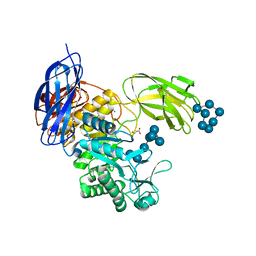 | | Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltoheptaose | | Descriptor: | CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, MALONATE ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Suzuki, R, Momma, M, Funane, K. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Isomaltooligosaccharide-binding structure ofPaenibacillussp. 598K cycloisomaltooligosaccharide glucanotransferase
Biosci. Rep., 37, 2017
|
|
5X7G
 
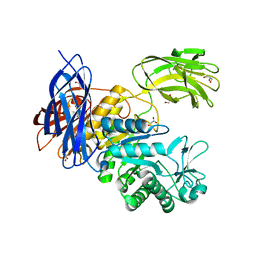 | | Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase | | Descriptor: | CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, GLYCEROL, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Suzuki, R, Momma, M, Funane, K. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isomaltooligosaccharide-binding structure ofPaenibacillussp. 598K cycloisomaltooligosaccharide glucanotransferase
Biosci. Rep., 37, 2017
|
|
2DX5
 
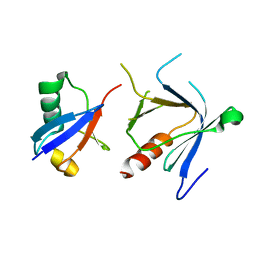 | | The complex structure between the mouse EAP45-GLUE domain and ubiquitin | | Descriptor: | Ubiquitin, Vacuolar protein sorting protein 36 | | Authors: | Hirano, S, Suzuki, N, Slagsvold, T, Kawasaki, M, Trambaiolo, D, Kato, R, Stenmark, H, Wakatsuki, S. | | Deposit date: | 2006-08-24 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain
Nat.Struct.Mol.Biol., 13, 2006
|
|
5X7O
 
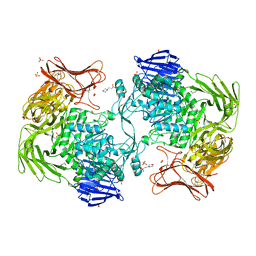 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fujimoto, Z, Suzuki, N, Kishine, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
5X7P
 
 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
5X7R
 
 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with isomaltohexaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
5X7S
 
 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase, terbium derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
5X7Q
 
 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with maltohexaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
3WNP
 
 | | D308A, F268V, D469Y, A513V, and Y515S quintuple mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoundecaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, R, Suzuki, N, Fujimoto, Z, Momma, M, Kimura, K, Kitamura, S, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular engineering of cycloisomaltooligosaccharide glucanotransferase from Bacillus circulans T-3040: structural determinants for the reaction product size and reactivity.
Biochem.J., 467, 2015
|
|
2ZVO
 
 | | NEMO CoZi domain in complex with diubiquitin in C2 space group | | Descriptor: | NF-kappa-B essential modulator, UBC protein | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
2ZVN
 
 | | NEMO CoZi domain incomplex with diubiquitin in P212121 space group | | Descriptor: | NF-kappa-B essential modulator, UBC protein | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
2ZFA
 
 | | Structure of Lactate Oxidase at pH4.5 from AEROCOCCUS VIRIDANS | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Lactate oxidase | | Authors: | Furuichi, M, Balasundaresan, D, Suzuki, N, Yoshida, Y, Minagawa, H, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
8XE7
 
 | |
4Z4K
 
 | |
4Z4M
 
 | |
