1S7D
 
 | |
1S12
 
 | | Crystal structure of TM1457 | | Descriptor: | ACETATE ION, hypothetical protein TM1457 | | Authors: | Shin, D.H, Lou, Y, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of TM1457 from Thermotoga maritima.
J.Struct.Biol., 152, 2005
|
|
1ECY
 
 | | PROTEASE INHIBITOR ECOTIN | | Descriptor: | ECOTIN, alpha-D-glucopyranose, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Shin, D.H, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure analyses of uncomplexed ecotin in two crystal forms: implications for its function and stability.
Protein Sci., 5, 1996
|
|
1ECZ
 
 | | PROTEASE INHIBITOR ECOTIN | | Descriptor: | ECOTIN, octyl beta-D-glucopyranoside | | Authors: | Shin, D.H, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure analyses of uncomplexed ecotin in two crystal forms: implications for its function and stability.
Protein Sci., 5, 1996
|
|
1JX7
 
 | | Crystal structure of ychN protein from E.coli | | Descriptor: | HYPOTHETICAL PROTEIN YCHN | | Authors: | Shin, D.H, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2001-09-05 | | Release date: | 2002-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a conserved hypothetical protein from Escherichia coli
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
1MZM
 
 | |
7WHC
 
 | | Crystal structure of SARS-CoV-2 3CLpro catalytic domain | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Shin, D.H, Jo, S.R. | | Deposit date: | 2021-12-30 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Dimerization Tendency of 3CLpros of Human Coronaviruses Based on the X-ray Crystal Structure of the Catalytic Domain of SARS-CoV-2 3CLpro.
Int J Mol Sci, 23, 2022
|
|
6JQ8
 
 | |
5JXD
 
 | | Crystal structure of murine Tnfaip8 C165S mutant | | Descriptor: | Tumor necrosis factor alpha-induced protein 8, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Park, J, Kim, M.S, Lee, D, Shin, D.H. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The Tnfaip8-PE complex is a novel upstream effector in the anti-autophagic action of insulin
Sci Rep, 7, 2017
|
|
4PNH
 
 | |
4TN5
 
 | |
4EJ0
 
 | |
4GQX
 
 | | Crystal structure of EIIA(NTR) from Burkholderia pseudomallei | | Descriptor: | PTS IIA-like nitrogen-regulatory protein PtsN | | Authors: | Kim, M.-S, Shin, D.H. | | Deposit date: | 2012-08-24 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New molecular interaction of IIA(Ntr) and HPr from Burkholderia pseudomallei identified by X-ray crystallography and docking studies
Proteins, 81, 2013
|
|
1T6S
 
 | | Crystal structure of a conserved hypothetical protein from Chlorobium tepidum | | Descriptor: | NITRATE ION, conserved hypothetical protein | | Authors: | Kim, J.S, Shin, D.H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of ScpB from Chlorobium tepidum, a protein involved in chromosome partitioning.
Proteins, 62, 2006
|
|
5Z0B
 
 | |
5XHW
 
 | | Crystal structure of HddC from Yersinia pseudotuberculosis | | Descriptor: | Putative 6-deoxy-D-mannoheptose pathway protein, SULFATE ION | | Authors: | Park, J, Kim, H, Kim, S, Shin, D.H. | | Deposit date: | 2017-04-24 | | Release date: | 2018-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of d-glycero-alpha-d-manno-heptose-1-phosphate guanylyltransferase from Yersinia pseudotuberculosis.
Biochim. Biophys. Acta, 1866, 2018
|
|
5X9Q
 
 | | Crystal structure of HldC from Burkholderia pseudomallei | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-03-08 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of D-glycero-Beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei.
Proteins, 86, 2018
|
|
1LQL
 
 | | Crystal structure of OsmC like protein from Mycoplasma pneumoniae | | Descriptor: | osmotical inducible protein C like family | | Authors: | Choi, I.-G, Shin, D.H, Brandsen, J, Jancarik, J, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-05-10 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of a stress inducible protein from Mycoplasma pneumoniae at 2.85 A resolution
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
5XF2
 
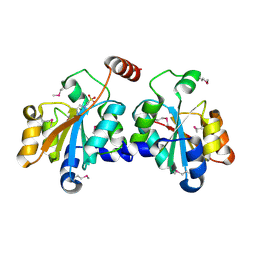 | | Crystal structure of SeMet-HldC from Burkholderia pseudomallei | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-04-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Expression and crystallographic studies of D-glycero-beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1NM3
 
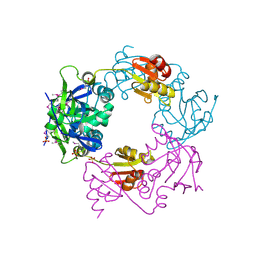 | | Crystal structure of Heamophilus influenza hybrid-Prx5 | | Descriptor: | Protein HI0572, SULFATE ION | | Authors: | Kim, S.J, Woo, J.R, Hwang, Y.S, Jeong, D.G, Shin, D.H, Kim, K.H, Ryu, S.E. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Tetrameric Structure of Haemophilus influenza Hybrid Prx5 Reveals Interactions between Electron Donor and Acceptor Proteins.
J.Biol.Chem., 278, 2003
|
|
2I1L
 
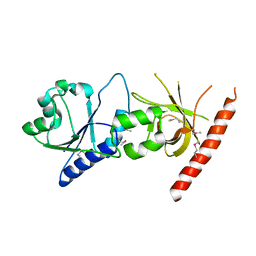 | | Crystal structure of the C2 form of FAD synthetase from Thermotoga maritima | | Descriptor: | Riboflavin kinase/FMN adenylyltransferase | | Authors: | Wang, W, Shin, D.H, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the C2 form of FAD synthetase from Thermotoga maritima
To be Published
|
|
1OIL
 
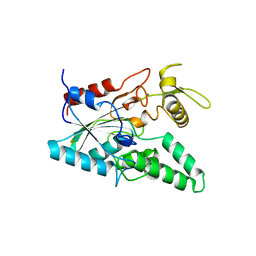 | | STRUCTURE OF LIPASE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Kim, K.K, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 1996-12-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor.
Structure, 5, 1997
|
|
1R5J
 
 | | Crystal Structure of a Phosphotransacetylase from Streptococcus pyogenes | | Descriptor: | putative phosphotransacetylase | | Authors: | Xu, Q.S, Shin, D.H, Pufan, R, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-10-10 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a phosphotransacetylase from Streptococcus pyogenes.
Proteins, 55, 2004
|
|
1OY5
 
 | | Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus | | Descriptor: | tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Liu, J, Wang, W, Shin, D.H, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-04-03 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus at 2.6 A resolution: a novel methyltransferase fold.
Proteins, 53, 2003
|
|
1RZL
 
 | | RICE NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, NONSPECIFIC LIPID TRANSFER PROTEIN, SULFATE ION | | Authors: | Lee, J.Y, Min, K.S, Cha, H, Shin, D.H, Hwang, K.Y, Suh, S.W. | | Deposit date: | 1997-10-09 | | Release date: | 1998-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rice non-specific lipid transfer protein: the 1.6 A crystal structure in the unliganded state reveals a small hydrophobic cavity.
J.Mol.Biol., 276, 1998
|
|
