3W67
 
 | | Crystal structure of mouse alpha-tocopherol transfer protein in complex with alpha-tocopherol and phosphatidylinositol-(3,4)-bisphosphate | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dibutanoate, Alpha-tocopherol transfer protein | | Authors: | Ohto, U, Satow, Y. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Impaired alpha-TTP-PIPs interaction underlies familial vitamin E deficiency
Science, 340, 2013
|
|
5SIC
 
 | |
3SSI
 
 | |
2SIC
 
 | | REFINED CRYSTAL STRUCTURE OF THE COMPLEX OF SUBTILISIN BPN' AND STREPTOMYCES SUBTILISIN INHIBITOR AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, STREPTOMYCES SUBTILISIN INHIBITOR (SSI), SUBTILISIN BPN' | | Authors: | Mitsui, Y, Takeuchi, Y, Hirono, S, Akagawa, H, Nakamura, K.T. | | Deposit date: | 1991-04-01 | | Release date: | 1993-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the complex of subtilisin BPN' and Streptomyces subtilisin inhibitor at 1.8 A resolution.
J.Mol.Biol., 221, 1991
|
|
3SIC
 
 | |
6IDG
 
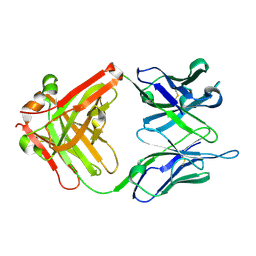 | | antibody 64M-5 Fab in complex with dT(6-4)T | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain), DNA (5'-D(*(64T)P*(5PY))-3') | | Authors: | Yokoyama, H, Mizutani, R, Noguchi, S, Hayashida, N. | | Deposit date: | 2018-09-10 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the antibody 64M-5 Fab and its complex with dT(6-4)T indicate induced-fit and high-affinity mechanisms.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6IDH
 
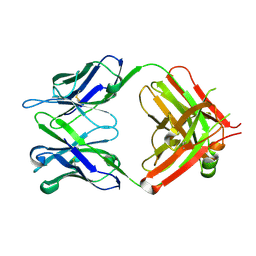 | | Antibody 64M-5 Fab in ligand-free form | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain) | | Authors: | Yokoyama, H, Mizutani, R, Noguchi, S, Hayashida, N. | | Deposit date: | 2018-09-10 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the antibody 64M-5 Fab and its complex with dT(6-4)T indicate induced-fit and high-affinity mechanisms.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
2TLD
 
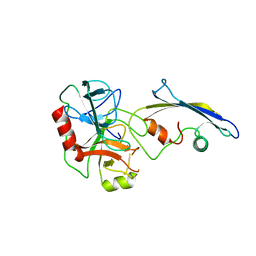 | | CRYSTAL STRUCTURE OF AN ENGINEERED SUBTILISIN INHIBITOR COMPLEXED WITH BOVINE TRYPSIN | | Descriptor: | STREPTOMYCES SUBTILISIN INHIBITOR (SSI), TRYPSIN | | Authors: | Mitsui, Y, Takeuchi, Y, Nonaka, T, Nakamura, K.T. | | Deposit date: | 1991-09-16 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an engineered subtilisin inhibitor complexed with bovine trypsin.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
2ILA
 
 | |
1GPH
 
 | |
1SRD
 
 | | Three-dimensional structure of CU,ZN-superoxide dismutase from spinach at 2.0 Angstroms resolution | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Kitagawa, Y, Katsube, Y. | | Deposit date: | 1993-04-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of Cu,Zn-superoxide dismutase from spinach at 2.0 A resolution.
J.Biochem.(Tokyo), 109, 1991
|
|
1KVB
 
 | | E. COLI RIBONUCLEASE HI D134H MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1MDA
 
 | | CRYSTAL STRUCTURE OF AN ELECTRON-TRANSFER COMPLEX BETWEEN METHYLAMINE DEHYDROGENASE AND AMICYANIN | | Descriptor: | AMICYANIN, COPPER (II) ION, METHYLAMINE DEHYDROGENASE (HEAVY SUBUNIT), ... | | Authors: | Chen, L, Durley, R, Mathews, F.S. | | Deposit date: | 1992-03-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an electron-transfer complex between methylamine dehydrogenase and amicyanin.
Biochemistry, 31, 1992
|
|
2MTA
 
 | | CRYSTAL STRUCTURE OF A TERNARY ELECTRON TRANSFER COMPLEX BETWEEN METHYLAMINE DEHYDROGENASE, AMICYANIN AND A C-TYPE CYTOCHROME | | Descriptor: | AMICYANIN, COPPER (II) ION, CYTOCHROME C551I, ... | | Authors: | Chen, L, Mathews, F.S. | | Deposit date: | 1993-10-26 | | Release date: | 1994-01-31 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an electron transfer complex: methylamine dehydrogenase, amicyanin, and cytochrome c551i.
Science, 264, 1994
|
|
3THC
 
 | |
3THD
 
 | | Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2011-08-18 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of human beta-galactosidase: structural basis of Gm1 gangliosidosis and morquio B diseases
J.Biol.Chem., 287, 2012
|
|
1KVA
 
 | | E. COLI RIBONUCLEASE HI D134A MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVC
 
 | | E. COLI RIBONUCLEASE HI D134N MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
