8UPY
 
 | | Methanosarcine mazei tRNAPyl in A-site of ribosome | | Descriptor: | RNA (72-MER) | | Authors: | Krahn, N, Zhang, J, Melnikov, S.V, Tharp, J.M, Villa, A, Patel, A, Howard, R.J, Gabir, H, Patel, T.R, Stetefeld, J, Puglisi, J, Soll, D. | | Deposit date: | 2023-10-23 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.
Nucleic Acids Res., 52, 2024
|
|
4V3L
 
 | | RNF38-UB-UbcH5B-Ub complex | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF38, POLYUBIQUITIN-C, ... | | Authors: | Buetow, L, Gabrielsen, M, Anthony, N.G, Dou, H, Patel, A, Aitkenhead, H, Sibbet, G.J, Smith, B.O, Huang, D.T. | | Deposit date: | 2014-10-20 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Activation of a Primed Ring E3-E2-Ubiquitin Complex by Non-Covalent Ubiquitin.
Mol.Cell, 58, 2015
|
|
4V3K
 
 | | RNF38-UbcH5B-UB complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE RNF38, ... | | Authors: | Buetow, L, Gabrielsen, M, Anthony, N.G, Dou, H, Patel, A, Aitkenhead, H, Sibbet, G.J, Smith, B.O, Huang, D.T. | | Deposit date: | 2014-10-20 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Activation of a Primed Ring E3-E2-Ubiquitin Complex by Non-Covalent Ubiquitin.
Mol.Cell, 58, 2015
|
|
8UPT
 
 | | Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome | | Descriptor: | RNA (71-MER) | | Authors: | Krahn, N, Zhang, J, Melnikov, S.V, Tharp, J.M, Villa, A, Patel, A, Howard, R.J, Gabir, H, Patel, T.R, Stetefeld, J, Puglisi, J, Soll, D. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.
Nucleic Acids Res., 52, 2024
|
|
2UUN
 
 | | Crystal structure of C-phycocyanin from Phormidium, Lyngbya spp. (Marine) and Spirulina sp. (Fresh water) shows two different ways of energy transfer between two hexamers. | | Descriptor: | BILIVERDINE IX ALPHA, C-PHYCOCYANIN, PHYCOCYANOBILIN | | Authors: | Satyanarayana, L, Patel, A, Mishra, S, K Ghosh, P, Suresh, C.G. | | Deposit date: | 2007-03-05 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of C-Phycocyanin from Phormidium, Lyngbya Spp. (Marine) and Spirulina Sp. (Fresh Water) Shows Two Different Ways of Energy Transfer between Two Hexamers.
To be Published
|
|
1F2E
 
 | | STRUCTURE OF SPHINGOMONAD, GLUTATHIONE S-TRANSFERASE COMPLEXED WITH GLUTATHIONE | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Nishio, T, Watanabe, T, Patel, A, Wang, Y, Lau, P.C.K, Grochulski, P, Li, Y, Cygler, M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-06-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Properties of a Sphingomonad and Marine Bacterium Beta-Class Glutathione S-Transferases and Crystal Structure of the Former Complex with Glutathione
To be published
|
|
8CX9
 
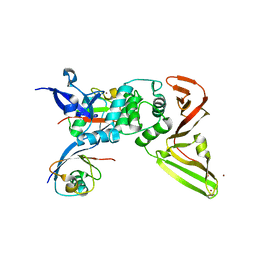 | | Structure of the SARS-COV2 PLpro (C111S) in complex with a dimeric Ubv that inhibits activity by an unusual allosteric mechanism | | Descriptor: | BROMIDE ION, CHLORIDE ION, Papain-like protease nsp3, ... | | Authors: | Singer, A.U, Slater, C.L, Patel, A, Russel, R, Mark, B.L, Sidhu, S.S. | | Deposit date: | 2022-05-20 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site.
Plos Pathog., 18, 2022
|
|
2UUM
 
 | | Crystal structure of C-phycocyanin from Phormidium, Lyngbya spp. (Marine) and Spirulina sp. (Fresh water) shows two different ways of energy transfer between two hexamers. | | Descriptor: | BILIVERDINE IX ALPHA, C-PHYCOCYANIN ALPHA CHAIN, C-PHYCOCYANIN BETA CHAIN, ... | | Authors: | Satyanarayana, L, Patel, A, Mishra, S, K Ghosh, P, Suresh, C.G. | | Deposit date: | 2007-03-04 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of C-Phycocyanin from Phormidium, Lyngbya Spp. (Marine) and Spirulina Sp. (Fresh Water) Shows Two Different Ways of Energy Transfer between Two Hexamers.
To be Published
|
|
8EYG
 
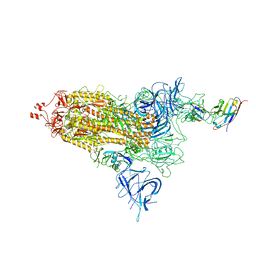 | |
8EYH
 
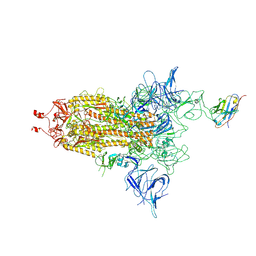 | | SARS-CoV-2 spike protein bound with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Laughlin, Z.T, Patel, A, Ortlund, E.A. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | SARS-CoV-2 spike protein bound with nanobody
To Be Published
|
|
8F4P
 
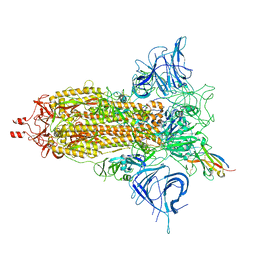 | |
2CSD
 
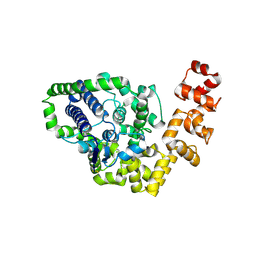 | |
2CSB
 
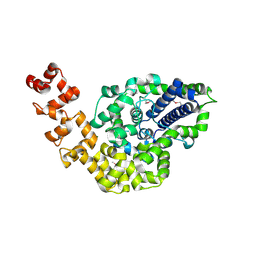 | | Crystal structure of Topoisomerase V from Methanopyrus kandleri (61 kDa fragment) | | Descriptor: | MAGNESIUM ION, Topoisomerase V | | Authors: | Taneja, B, Patel, A, Slesarev, A, Mondragon, A. | | Deposit date: | 2005-05-21 | | Release date: | 2006-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases
Embo J., 25, 2006
|
|
4X4J
 
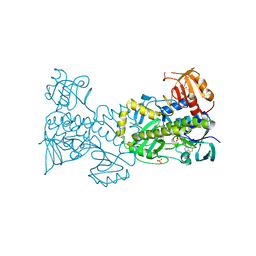 | | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative oxygenase, SULFATE ION | | Authors: | Tsai, S.-C, Jackson, D.R, Patel, A, Barajas, J.F, Rohr, J, Yu, X, Liu, H.-W, Sasaki, E, Calveras, J, Metsa-Ketela, M. | | Deposit date: | 2014-12-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis
To Be Published
|
|
4XS7
 
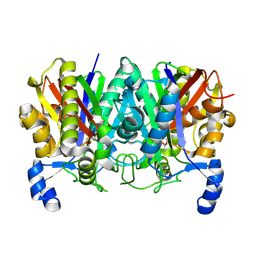 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XSA
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XSB
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XS9
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase, N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-[2-(propanoylamino)ethyl]-beta-alaninamide | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
5TT4
 
 | | Determining the Molecular Basis For Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Patel, A, Tsai, S.C, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2016-11-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determining the Molecular Basis For Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4ES0
 
 | | X-ray structure of WDR5-SETd1b Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase SETD1B, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.817 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERY
 
 | | X-ray structure of WDR5-MLL3 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERQ
 
 | | X-ray structure of WDR5-MLL2 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL2, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4EYT
 
 | | Crystal structure of the C-terminal domain of Tetrahymena telomerase protein p65 | | Descriptor: | SULFATE ION, Telomerase associated protein p65 | | Authors: | Singh, M, Wang, Z, Koo, B.-K, Patel, A, Cascio, D, Collins, K, Feigon, J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Telomerase RNA Recognition and RNP Assembly by the Holoenzyme La Family Protein p65.
Mol.Cell, 47, 2012
|
|
4ESG
 
 | | X-ray structure of WDR5-MLL1 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERD
 
 | | Crystal structure of the C-terminal domain of Tetrahymena telomerase protein p65 in complex with stem IV of telomerase RNA | | Descriptor: | 5'-R(P*GP*GP*UP*CP*GP*AP*CP*AP*UP*CP*UP*UP*CP*GP*GP*AP*UP*GP*GP*AP*CP*C)-3', POTASSIUM ION, Telomerase associated protein p65 | | Authors: | Singh, M, Wang, Z, Koo, B.-K, Patel, A, Cascio, D, Collins, K, Feigon, J. | | Deposit date: | 2012-04-19 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.589 Å) | | Cite: | Structural Basis for Telomerase RNA Recognition and RNP Assembly by the Holoenzyme La Family Protein p65.
Mol.Cell, 47, 2012
|
|
