7ZK4
 
 | | The ABCB1 L335C mutant (mABCB1) in the outward facing state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZKB
 
 | | ABCB1 V978C mutant (mABCB1) in the inward facing state | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1 | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK8
 
 | | ABCB1 L971C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
6HIF
 
 | | Kuenenia stuttgartiensis hydrazine dehydrogenase complex | | Descriptor: | GLYCEROL, HEME C, Hydrazine dehydrogenase, ... | | Authors: | Akram, M, Dietl, A, Mersdorf, U, Prinz, S, Maalcke, W, Keltjens, J, Ferousi, C, de Almeida, N.M, Reimann, J, Kartal, B, Jetten, M.S.M, Parey, K, Barends, T.R.M. | | Deposit date: | 2018-08-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A 192-heme electron transfer network in the hydrazine dehydrogenase complex.
Sci Adv, 5, 2019
|
|
8POK
 
 | | Cryo-EM structure of cell-free synthesized human histamine H2 receptor coupled to heterotrimeric Gs protein in lipid environment | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, HISTAMINE, ... | | Authors: | Schnelle, K, Koeck, Z, Persechino, M, Umbach, S, Schihada, H, Januliene, D, Parey, K, Pockes, S, Kolb, P, Doetsch, V, Moeller, A, Hilger, D, Bernhard, F. | | Deposit date: | 2023-07-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of cell-free synthesized human histamine 2 receptor/G s complex in nanodisc environment.
Nat Commun, 15, 2024
|
|
8C80
 
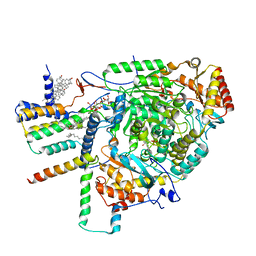 | | Cryo-EM structure of the yeast SPT-Orm1-Monomer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ERGOSTEROL, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
8C81
 
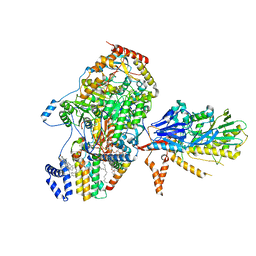 | | Cryo-EM structure of the yeast SPT-Orm1-Sac1 complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ERGOSTEROL, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
8C82
 
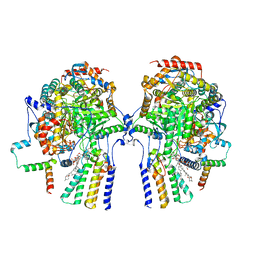 | | Cryo-EM structure of the yeast SPT-Orm1-Dimer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
7PH2
 
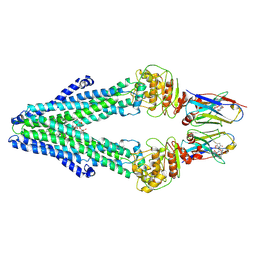 | | Nanodisc reconstituted MsbA in complex with nanobodies, spin-labeled at position A60C | | Descriptor: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,5-bis(oxidanyl)oxan-2-yl]oxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{R})-3-nonanoyloxytetradecanoyl]oxy-5-[[(3~{R})-3-octanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{S},5~{S},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanylnonanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-oxan-2-yl]methoxy]-5-oxidanyl-oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase, ... | | Authors: | Januliene, D, Parey, K, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
3MMC
 
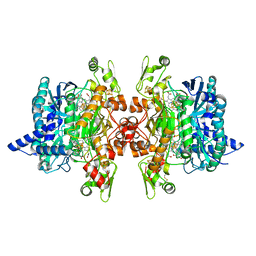 | | Structure of the dissimilatory sulfite reductase from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Schiffer, A, Parey, K, Warkentin, E, Diederichs, K, Huber, H, Stetter, K.O, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the dissimilatory sulfite reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Mol.Biol., 379, 2008
|
|
9FV0
 
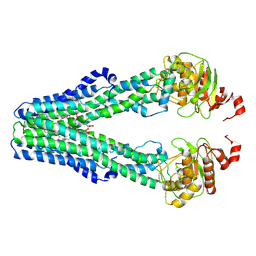 | | MsbA in MSP1D1 Nanodisc inward-facing narrow open | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shearer, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FUS
 
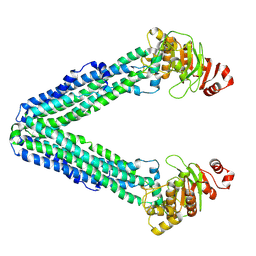 | | MsbA in LMNG inward-facing wide open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FV4
 
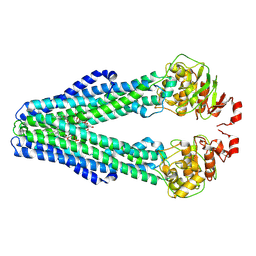 | | MsbA in peptidisc inward-facing narrow (-) open | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FV5
 
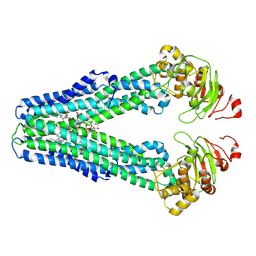 | | MsbA in peptidisc inward-facing narrow open | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FV8
 
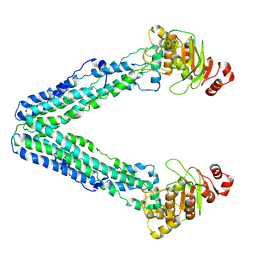 | | MsbA in Amphipol 18 inward-facing wide open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FUR
 
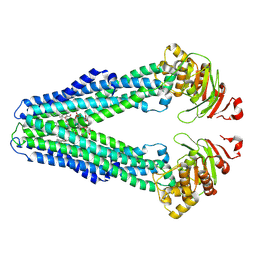 | | MsbA in LMNG inward-facing narrow | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FUW
 
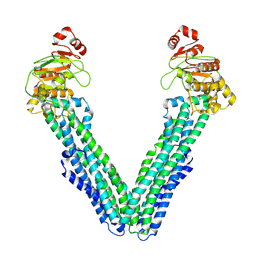 | | MsbA in Triton inward-facing wide (+) open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FUZ
 
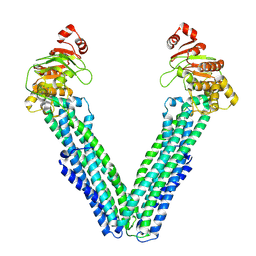 | | MsbA in UDM inward-facing wide (+) open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FV3
 
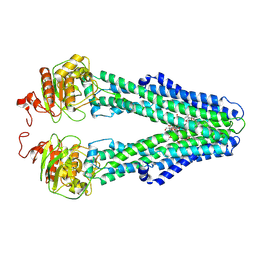 | | MsbA in MSP2N2 Nanodisc inward-facing narrow open | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FUU
 
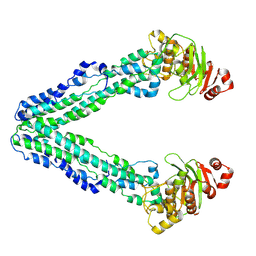 | | MsbA in GDN inward-facing wide open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FUY
 
 | | MsbA in UDM inward-facing wide open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FV7
 
 | | MsbA in Amphipol A8-35 inward-facing wide (+) open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FUQ
 
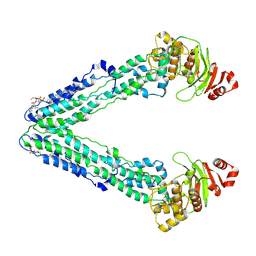 | | MsbA in DDM inward-facing wide open | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FV9
 
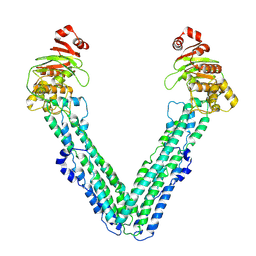 | | MsbA in Amphipol 18 inward-facing wide (+) open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
9FV6
 
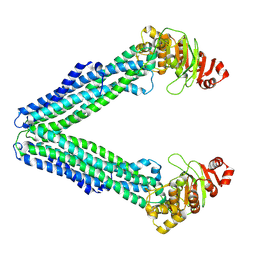 | | MsbA in Amphipol A8-35 inward-facing wide open | | Descriptor: | ATP-dependent lipid A-core flippase | | Authors: | Hoffmann, L, Baier, A, Jorde, L, Kamel, M, Schaefer, J, Schnelle, K, Scholz, A, Shvarev, D, Wong, J, Parey, K, Januliene, D, Moeller, A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The ABC transporter MsbA in a dozen environments.
Structure, 33, 2025
|
|
