1WQ8
 
 | | Crystal structure of Vammin, a VEGF-F from a snake venom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Vascular endothelial growth factor toxin | | Authors: | Suto, K, Yamazaki, Y, Morita, T, Mizuno, H. | | Deposit date: | 2004-09-23 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of novel vascular endothelial growth factors (VEGF) from snake venoms: insight into selective VEGF binding to kinase insert domain-containing receptor but not to fms-like tyrosine kinase-1.
J.Biol.Chem., 280, 2005
|
|
1WQ9
 
 | | Crystal structure of VR-1, a VEGF-F from a snake venom | | Descriptor: | Vascular endothelial growth factor | | Authors: | Suto, K, Yamazaki, Y, Morita, T, Mizuno, H. | | Deposit date: | 2004-09-24 | | Release date: | 2004-12-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of novel vascular endothelial growth factors (VEGF) from snake venoms: insight into selective VEGF binding to kinase insert domain-containing receptor but not to fms-like tyrosine kinase-1.
J.Biol.Chem., 280, 2005
|
|
8IDQ
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus with xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
8IDP
 
 | | Crystal structure of reducing-end xylose-releasing exoxylanase in GH30 from Talaromyces cellulolyticus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamichi, Y, Watanabe, M, Fujii, T, Inoue, H, Morita, T. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of reducing-end xylose-releasing exoxylanase in subfamily 7 of glycoside hydrolase family 30.
Proteins, 91, 2023
|
|
8JOR
 
 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type A crystal | | Descriptor: | Acyltransferase, PENTAETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
8JOS
 
 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type B crystal | | Descriptor: | Acyltransferase, CHLORIDE ION, TRIETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
1JWI
 
 | | Crystal Structure of Bitiscetin, a von Willeband Factor-dependent Platelet Aggregation Inducer. | | Descriptor: | bitiscetin, platelet aggregation inducer | | Authors: | Hirotsu, S, Mizuno, H, Fukuda, K, Qi, M.C, Matsui, T, Hamako, J, Morita, T, Titani, K. | | Deposit date: | 2001-09-04 | | Release date: | 2001-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of bitiscetin, a von Willebrand factor-dependent platelet aggregation inducer.
Biochemistry, 40, 2001
|
|
1C3A
 
 | | CRYSTAL STRUCTURE OF FLAVOCETIN-A FROM THE HABU SNAKE VENOM, A NOVEL CYCLIC TETRAMER OF C-TYPE LECTIN-LIKE HETERODIMERS | | Descriptor: | FLAVOCETIN-A: ALPHA SUBUNIT, FLAVOCETIN-A: BETA SUBUNIT | | Authors: | Fukuda, K, Mizuno, H, Atoda, H, Morita, T. | | Deposit date: | 1999-07-27 | | Release date: | 2000-03-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of flavocetin-A, a platelet glycoprotein Ib-binding protein, reveals a novel cyclic tetramer of C-type lectin-like heterodimers.
Biochemistry, 39, 2000
|
|
1IOD
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN THE COAGULATION FACTOR X BINDING PROTEIN FROM SNAKE VENOM AND THE GLA DOMAIN OF FACTOR X | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X BINDING PROTEIN, COAGULATION FACTOR X GLA DOMAIN | | Authors: | Mizuno, H, Fujimoto, Z, Atoda, H, Morita, T. | | Deposit date: | 2001-02-27 | | Release date: | 2001-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an anticoagulant protein in complex with the Gla domain of factor X.
Proc.Natl.Acad.Sci.Usa, 98, 2001
|
|
1X2W
 
 | | Crystal Structure of Apo-Habu IX-bp at pH 4.6 | | Descriptor: | CHLORIDE ION, Coagulation factor IX/X-binding protein A chain, Coagulation factor IX/factor X-binding protein B chain, ... | | Authors: | Suzuki, N, Fujimoto, Z, Morita, T, Fukamizu, A, Mizuno, H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | pH-Dependent Structural Changes at Ca(2+)-binding sites of Coagulation Factor IX-binding Protein
J.Mol.Biol., 353, 2005
|
|
1X2T
 
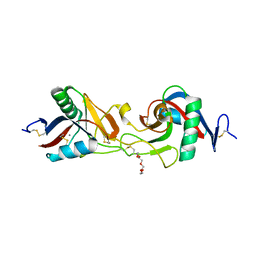 | | Crystal Structure of Habu IX-bp at pH 6.5 | | Descriptor: | CALCIUM ION, Coagulation factor IX/X-binding protein A chain, Coagulation factor IX/factor X-binding protein B chain, ... | | Authors: | Suzuki, N, Fujimoto, Z, Morita, T, Fukamizu, A, Mizuno, H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | pH-Dependent Structural Changes at Ca(2+)-binding sites of Coagulation Factor IX-binding Protein
J.Mol.Biol., 353, 2005
|
|
2DDB
 
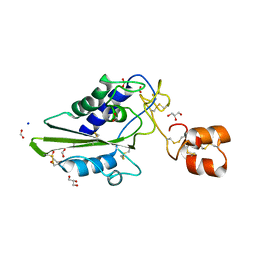 | | Crystal structure of pseudecin from Pseudechis porphyriacus | | Descriptor: | FORMIC ACID, GLYCEROL, Pseudecin, ... | | Authors: | Suzuki, N, Yamazaki, Y, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2006-01-25 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of pseudechetoxin and pseudecin, two snake-venom cysteine-rich secretory proteins that target cyclic nucleotide-gated ion channels: implications for movement of the C-terminal cysteine-rich domain
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2DDA
 
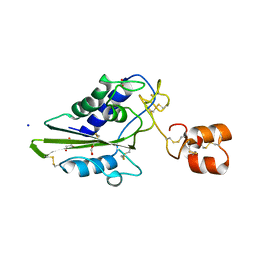 | | Crystal structure of pseudechetoxin from Pseudechis australis | | Descriptor: | FORMIC ACID, GLYCEROL, Pseudechetoxin, ... | | Authors: | Suzuki, N, Yamazaki, Y, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2006-01-25 | | Release date: | 2007-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of pseudechetoxin and pseudecin, two snake-venom cysteine-rich secretory proteins that target cyclic nucleotide-gated ion channels: implications for movement of the C-terminal cysteine-rich domain
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2EPF
 
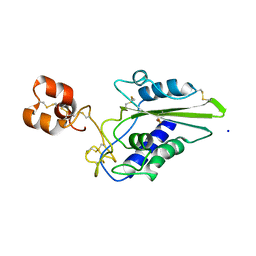 | | Crystal Structure of Zinc-Bound Pseudecin From Pseudechis Porphyriacus | | Descriptor: | Pseudecin, SODIUM ION, ZINC ION | | Authors: | Suzuki, N, Yamazaki, Y, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2007-03-29 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of pseudechetoxin and pseudecin, two snake-venom cysteine-rich secretory proteins that target cyclic nucleotide-gated ion channels: implications for movement of the C-terminal cysteine-rich domain
Acta Crystallogr.,Sect.D, 64, 2008
|
|
8WOP
 
 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, wild-type | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase 2, URIDINE-5'-DIPHOSPHATE | | Authors: | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | Deposit date: | 2023-10-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8WOV
 
 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, G233A mutant | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase 2, URIDINE-5'-DIPHOSPHATE | | Authors: | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | Deposit date: | 2023-10-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8WOW
 
 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, I160L/G233A mutant | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, UDP-glucose 4-epimerase 2, ... | | Authors: | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | Deposit date: | 2023-10-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
7C3N
 
 | | Crystal structure of JAK3 in complex with Delgocitinib | | Descriptor: | 3-[(3S,4R)-3-methyl-7-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1,7-diazaspiro[3.4]octan-1-yl]-3-oxidanylidene-propanenitrile, Tyrosine-protein kinase JAK3 | | Authors: | Doi, S, Otira, T, Kikuwaka, M, Nomura, A, Noji, S, Adachi, T. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a Janus Kinase Inhibitor Bearing a Highly Three-Dimensional Spiro Scaffold: JTE-052 (Delgocitinib) as a New Dermatological Agent to Treat Inflammatory Skin Disorders.
J.Med.Chem., 63, 2020
|
|
2DXS
 
 | | Crystal structure of HCV NS5B RNA polymerase complexed with a tetracyclic inhibitor | | Descriptor: | Genome polyprotein, N-[(13-CYCLOHEXYL-6,7-DIHYDROINDOLO[1,2-D][1,4]BENZOXAZEPIN-10-YL)CARBONYL]-2-METHYL-L-ALANINE | | Authors: | Adachi, T, Tsuruha, J, Doi, S, Murase, K, Ikegashira, K, Watanabe, S, Uehara, K, Orita, T, Nomura, A, Kamada, M. | | Deposit date: | 2006-08-30 | | Release date: | 2006-12-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Conformationally Constrained Tetracyclic Compounds as Potent Hepatitis C Virus NS5B RNA Polymerase Inhibitors
J.Med.Chem., 49, 2006
|
|
3VQS
 
 | | Crystal structure of HCV NS5B RNA polymerase with a novel piperazine inhibitor | | Descriptor: | (2R)-4-(5-cyclopropyl[1,3]thiazolo[4,5-d]pyrimidin-2-yl)-N-[3-fluoro-4-(trifluoromethoxy)benzyl]-1-{[4-(trifluoromethyl)phenyl]sulfonyl}piperazine-2-carboxamide, CHLORIDE ION, RNA-directed RNA polymerase | | Authors: | Adachi, T, Doi, S, Ando, I, Sugimoto, K, Orita, T, Nomura, A, Kamada, M. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Preclinical Characterization of JTK-853, a Novel Nonnucleoside Inhibitor of the Hepatitis C Virus RNA-Dependent RNA Polymerase.
Antimicrob.Agents Chemother., 56, 2012
|
|
8OQC
 
 | | Dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (1 h soaking) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Ribonuclease pancreatic, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
8OQG
 
 | | Cross-linked crystal of dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (high temperature data collection) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Ribonuclease pancreatic, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
8OQD
 
 | | Dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (1 h soaking) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Ribonuclease pancreatic, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
8OQE
 
 | | Dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (6 h soaking) | | Descriptor: | CHLORIDE ION, FORMIC ACID, Ribonuclease pancreatic, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
8OQF
 
 | | Cross-linked crystal of Dirhodium tetraacetate/ribonuclease A adduct in the P3221 space group (low temperature data collection) | | Descriptor: | (mi2-acetato-O, O')-hexaaquo-dirhodium (II), CHLORIDE ION, ... | | Authors: | Loreto, D, Merlino, A, Maity, B, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cross-Linked Crystals of Dirhodium Tetraacetate/RNase A Adduct Can Be Used as Heterogeneous Catalysts.
Inorg.Chem., 62, 2023
|
|
