1C9H
 
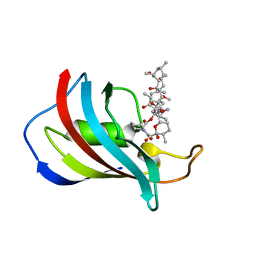 | | CRYSTAL STRUCTURE OF FKBP12.6 IN COMPLEX WITH RAPAMYCIN | | Descriptor: | FKBP12.6, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Deivanayagam, C.C.S, Carson, M, Thotakura, A, Narayana, S.V.L, Chodavarapu, C.S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of FKBP12.6 in complex with rapamycin.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4JCN
 
 | |
2ODQ
 
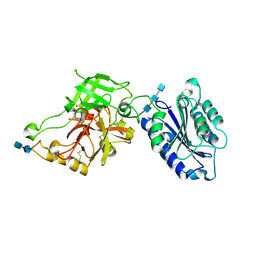 | | Complement component C2a, the catalytic fragment of C3- and C5-convertase of human complement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Narayana, S.V.L, Krishnan, V. | | Deposit date: | 2006-12-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of c2a, the catalytic fragment of classical pathway c3 and c5 convertase of human complement.
J.Mol.Biol., 367, 2007
|
|
2ODP
 
 | | Complement component C2a, the catalytic fragment of C3- and C5-convertase of human complement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Narayana, S.V.L, Krishnan, V. | | Deposit date: | 2006-12-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of c2a, the catalytic fragment of classical pathway c3 and c5 convertase of human complement.
J.Mol.Biol., 367, 2007
|
|
3PHS
 
 | |
3QDH
 
 | |
3RBI
 
 | |
3RBJ
 
 | |
3RCC
 
 | |
3RBK
 
 | |
3TVY
 
 | |
3TXA
 
 | | Structural Analysis of Adhesive Tip pilin, GBS104 from Group B Streptococcus agalactiae | | Descriptor: | CADMIUM ION, Cell wall surface anchor family protein, LITHIUM ION, ... | | Authors: | Krishnan, V, Narayana, S.V.L. | | Deposit date: | 2011-09-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Structure of Streptococcus agalactiae tip pilin GBS104: a model for GBS pili assembly and host interactions.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3TW0
 
 | |
3FRP
 
 | | Crystal Structure of Cobra Venom Factor, a Co-factor for C3- and C5 convertase CVFBb | | Descriptor: | CALCIUM ION, Cobra venom factor alpha chain, Cobra venom factor beta chain, ... | | Authors: | Krishnan, V, Narayana, S.V.L. | | Deposit date: | 2009-01-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The Crystal Structure of Cobra Venom Factor, a Cofactor for C3- and C5-Convertase CVFBb.
Structure, 17, 2009
|
|
2RAL
 
 | | Crystal Structure Analysis of double cysteine mutant of S.epidermidis adhesin SdrG: Evidence for the Dock,Lock and Latch ligand binding mechanism | | Descriptor: | Serine-aspartate repeat-containing protein G | | Authors: | Ponnuraj, K, Sthanam, N, Bowden, M.G, Hook, M. | | Deposit date: | 2007-09-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for the "dock, lock, and latch" ligand binding mechanism of the staphylococcal microbial surface component recognizing adhesive matrix molecules (MSCRAMM) SdrG.
J.Biol.Chem., 283, 2008
|
|
6POO
 
 | |
3TB7
 
 | |
3TBE
 
 | |
3ERB
 
 | |
1DF0
 
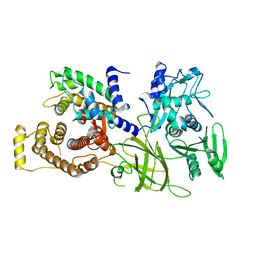 | | Crystal structure of M-Calpain | | Descriptor: | CALPAIN, M-CALPAIN | | Authors: | Hosfield, C.M, Elce, J.S, Davies, P.L, Jia, Z. | | Deposit date: | 1999-11-16 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of calpain reveals the structural basis for Ca(2+)-dependent protease activity and a novel mode of enzyme activation.
EMBO J., 18, 1999
|
|
3DF0
 
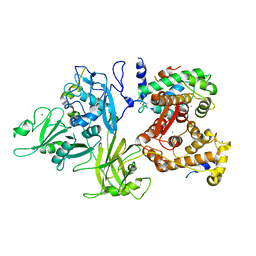 | | Calcium-dependent complex between m-calpain and calpastatin | | Descriptor: | CALCIUM ION, Calpain small subunit 1, Calpain-2 catalytic subunit, ... | | Authors: | Moldoveanu, T, Gehring, K, Green, D.R. | | Deposit date: | 2008-06-11 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains.
Nature, 456, 2008
|
|
