3TXA
 
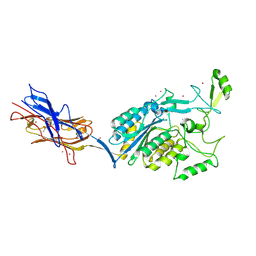 | | Structural Analysis of Adhesive Tip pilin, GBS104 from Group B Streptococcus agalactiae | | Descriptor: | CADMIUM ION, Cell wall surface anchor family protein, LITHIUM ION, ... | | Authors: | Krishnan, V, Narayana, S.V.L. | | Deposit date: | 2011-09-23 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Structure of Streptococcus agalactiae tip pilin GBS104: a model for GBS pili assembly and host interactions.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3TW0
 
 | |
3TVY
 
 | |
4K4F
 
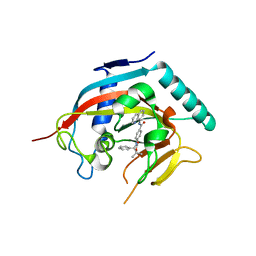 | | Co-crystal structure of TNKS1 with compound 18 [4-[(4S)-5,5-dimethyl-2-oxo-4-phenyl-1,3-oxazolidin-3-yl]-N-(quinolin-8-yl)benzamide] | | Descriptor: | 4-[(4S)-5,5-dimethyl-2-oxo-4-phenyl-1,3-oxazolidin-3-yl]-N-(quinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X, Bregman, H, Wilson, C, DiMauro, E, Gunaydin, H. | | Deposit date: | 2013-04-12 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of novel, induced-pocket binding oxazolidinones as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
4K4E
 
 | | Co-crystal structure of tnks1 with compound 52 [N~2-(5-chloro-2-methoxyphenyl)-N-[trans-4-(2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)cyclohexyl]glycinamide] | | Descriptor: | N~2~-(5-chloro-2-methoxyphenyl)-N-[trans-4-(2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)cyclohexyl]glycinamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-04-12 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of novel, induced-pocket binding oxazolidinones as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
3ENT
 
 | |
3ENU
 
 | |
3I9H
 
 | |
3HZ2
 
 | |
3HZB
 
 | |
3IAJ
 
 | |
4JL9
 
 | | Crystal structure of mouse TBK1 bound to BX795 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Li, P, Shu, C. | | Deposit date: | 2013-03-12 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.0999 Å) | | Cite: | Structural Insights into the Functions of TBK1 in Innate Antimicrobial Immunity.
Structure, 21, 2013
|
|
4JLC
 
 | | Crystal structure of mouse TBK1 bound to SU6668 | | Descriptor: | 3-{2,4-dimethyl-5-[(Z)-(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-1H-pyrrol-3-yl}propanoic acid, Serine/threonine-protein kinase TBK1 | | Authors: | Li, P. | | Deposit date: | 2013-03-12 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the Functions of TBK1 in Innate Antimicrobial Immunity.
Structure, 21, 2013
|
|
9G0L
 
 | |
