3IKK
 
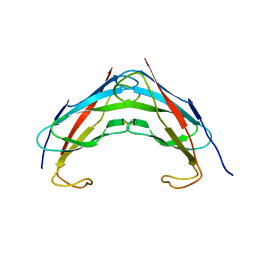 | | Crystal structure analysis of msp domain | | Descriptor: | Vesicle-associated membrane protein-associated protein B/C | | Authors: | Shi, J, Lua, S, Song, J. | | Deposit date: | 2009-08-06 | | Release date: | 2010-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elimination of the native structure and solubility of the hVAPB MSP domain by the Pro56Ser mutation that causes amyotrophic lateral sclerosis.
Biochemistry, 49, 2010
|
|
6MD6
 
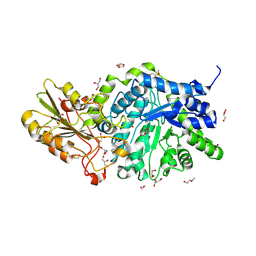 | | CRYSTAL STRUCTURE ANALYSIS OF PLANT EXOHYDROLASE IN COMPLEX WITH METHYL 2-THIO-BETA-SOPHOROSIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL, ... | | Authors: | Streltsov, V.A, Luang, S, Hrmova, M. | | Deposit date: | 2018-09-04 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
6MI1
 
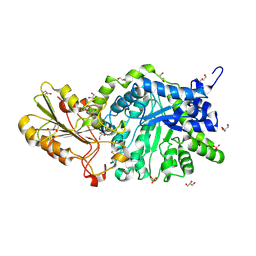 | | CRYSTAL STRUCTURE ANALYSIS OF THE VARIANT PLANT EXOHYDROLASE ARG158ALA-GLU161ALA IN COMPLEX WITH METHYL 6-THIO-BETA-GENTIOBIOSIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL, ... | | Authors: | Streltsov, V.A, Luang, S, Hrmova, M. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLQ
 
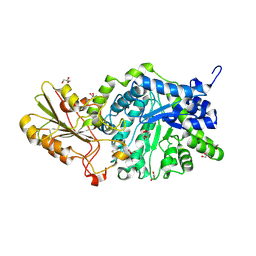 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL | | Authors: | Streltsov, V.A, Luang, S, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLR
 
 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL | | Authors: | Streltsov, V.A, Hrmova, M, Luang, S. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLO
 
 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, SULFATE ION, ... | | Authors: | Streltsov, V.A, Luang, S, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLP
 
 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 1-thio-beta-D-glucopyranose-(1-6)-methyl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | Authors: | Streltsov, V.A, Luang, S, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
4JIE
 
 | | Structural analysis and insights into glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-mannosidase/beta-glucosidase, GLYCEROL, ... | | Authors: | Tankrathok, A, Luang, S, Robinson, R.C, Kimura, A, Hrmova, M, Ketudat Cairns, J.R. | | Deposit date: | 2013-03-05 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis and insights into the glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JHO
 
 | | Structural analysis and insights into glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-mannosidase/beta-glucosidase, GLYCEROL | | Authors: | Tankrathok, A, Luang, S, Robinson, R.C, Kimura, A, Hrmova, M, Ketudat Cairns, J.R. | | Deposit date: | 2013-03-05 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural analysis and insights into the glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1Y0O
 
 | |
1U79
 
 | | Crystal structure of AtFKBP13 | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase 3 | | Authors: | Gopalan, G, Swaminathan, K. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis uncovers a role for redox in regulating FKBP13, an immunophilin of the chloroplast thylakoid lumen
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
7F2J
 
 | | Crystal structure of AtFKBP53 FKBD in complex with rapamycin | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase FKBP53, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Singh, A.K, Saharan, K, Vasudevan, D. | | Deposit date: | 2021-06-11 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal packing reveals rapamycin-mediated homodimerization of an FK506-binding domain.
Int.J.Biol.Macromol., 206, 2022
|
|
3RFY
 
 | |
3WLJ
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with 3-deoxy-glucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLH
 
 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLL
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with PEG400 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[beta-D-xylopyranose-(1-2)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLN
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with octyl-S-glucoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLI
 
 | | Crystal Structure Analysis of Plant Exohydrolase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL, ... | | Authors: | Streltsov, V.A, Hrmova, M. | | Deposit date: | 2013-11-12 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
3WLM
 
 | |
3WLK
 
 | |
6LKB
 
 | | Crystal Structure of the peptidylprolyl isomerase domain of Arabidopsis thaliana CYP71. | | Descriptor: | COBALT (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lakhanpal, S, Jobichen, C, Swaminathan, K. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analyses of the PPIase domain of Arabidopsis thaliana CYP71 reveal its catalytic activity toward histone H3.
Febs Lett., 595, 2021
|
|
9CC8
 
 | | Hexameric state of the NRC4 resistosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NLR-required for cell death 4 | | Authors: | Liu, F, Yang, Z, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2024-06-21 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Activation of the helper NRC4 immune receptor forms a hexameric resistosome.
Cell, 187, 2024
|
|
9CC9
 
 | | Dodecameric state of the NRC4 resistosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NLR-required for cell death 4 | | Authors: | Liu, F, Yang, Z, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2024-06-21 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Activation of the helper NRC4 immune receptor forms a hexameric resistosome.
Cell, 187, 2024
|
|
6J2M
 
 | | Crystal structure of AtFKBP53 C-terminal domain | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP53 | | Authors: | Singh, A.K, Vasudevan, D. | | Deposit date: | 2019-01-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | AtFKBP53: a chimeric histone chaperone with functional nucleoplasmin and PPIase domains.
Nucleic Acids Res., 48, 2020
|
|
6J2Z
 
 | | AtFKBP53 N-terminal Nucleoplasmin Domain | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP53 | | Authors: | Singh, A.K, Vasudevan, D. | | Deposit date: | 2019-01-03 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AtFKBP53: a chimeric histone chaperone with functional nucleoplasmin and PPIase domains.
Nucleic Acids Res., 48, 2020
|
|
