2Z27
 
 | | Thr109Ser dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
2EG6
 
 | |
2EG8
 
 | | The crystal structure of E. coli dihydroorotase complexed with 5-fluoroorotic acid | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-02-28 | | Release date: | 2007-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Ligand-free and Inhibitor Complexes of Dihydroorotase from Escherichia coli: Implications for Loop Movement in Inhibitor Design
J.Mol.Biol., 370, 2007
|
|
6XFM
 
 | | Molecular structure of the core of amyloid-like fibrils formed by residues 111-214 of FUS | | Descriptor: | RNA-binding protein FUS | | Authors: | Tycko, R, Lee, M, Ghosh, U, Thurber, K, Kato, M. | | Deposit date: | 2020-06-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Molecular structure and interactions within amyloid-like fibrils formed by a low-complexity protein sequence from FUS.
Nat Commun, 11, 2020
|
|
9CP7
 
 | |
9CP9
 
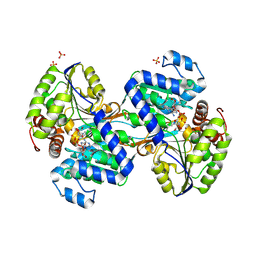 | | Crystal structure of DHPS-3-dehydrogenase, HpsN H319A variant from Cupriavidus pinatubonensis in complex with substrate (R-DHPS) and NADH | | Descriptor: | (2R)-2,3-dihydroxypropane-1-sulfonic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Lee, M. | | Deposit date: | 2024-07-18 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Structural and kinetic insights into the stereospecific oxidation of R -2,3-dihydroxypropanesulfonate by DHPS-3-dehydrogenase from Cupriavidus pinatubonensis.
Chem Sci, 15, 2024
|
|
9CP8
 
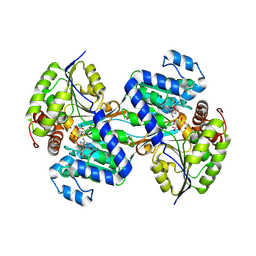 | |
6OWJ
 
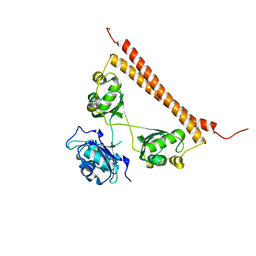 | | Zn-mediated polymerization of human SFPQ | | Descriptor: | Splicing factor, proline- and glutamine-rich, ZINC ION | | Authors: | Lee, M. | | Deposit date: | 2019-05-10 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis of the zinc-induced cytoplasmic aggregation of the RNA-binding protein SFPQ.
Nucleic Acids Res., 48, 2020
|
|
7SP0
 
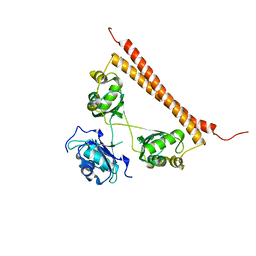 | | Crystal structure of human SFPQ L534I mutant in complex with zinc | | Descriptor: | Splicing factor, proline- and glutamine-rich, ZINC ION | | Authors: | Lee, M. | | Deposit date: | 2021-11-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Familial ALS-associated SFPQ variants promote the formation of SFPQ cytoplasmic aggregates in primary neurons.
Open Biology, 12, 2022
|
|
8EZE
 
 | |
8V35
 
 | | Crystal structure of HpsN from Cupriavidus pinatubonensis | | Descriptor: | 1,2-ETHANEDIOL, Sulfopropanediol 3-dehydrogenase, ZINC ION | | Authors: | Lee, M. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and kinetic insights into the stereospecific oxidation of R -2,3-dihydroxypropanesulfonate by DHPS-3-dehydrogenase from Cupriavidus pinatubonensis.
Chem Sci, 15, 2024
|
|
8V37
 
 | |
8V36
 
 | |
5K3X
 
 | | Crystal Structure of the sulfite dehydrogenase, SorT R78K mutant from Sinorhizobium meliloti | | Descriptor: | (MOLYBDOPTERIN-S,S)-OXO-MOLYBDENUM, GLYCEROL, Putative sulfite oxidase | | Authors: | Lee, M, McGrath, A, Maher, M. | | Deposit date: | 2016-05-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The central active site arginine in sulfite oxidizing enzymes alters kinetic properties by controlling electron transfer and redox interactions.
Biochim. Biophys. Acta, 1859, 2017
|
|
6VVI
 
 | |
6VVH
 
 | |
8TNF
 
 | |
8EZD
 
 | |
6NCQ
 
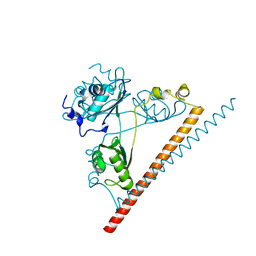 | | The dimerization domain of human SFPQ in space group C2221 | | Descriptor: | Splicing factor, proline- and glutamine-rich | | Authors: | Lee, M. | | Deposit date: | 2018-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new crystal structure and small-angle X-ray scattering analysis of the homodimer of human SFPQ.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7JGW
 
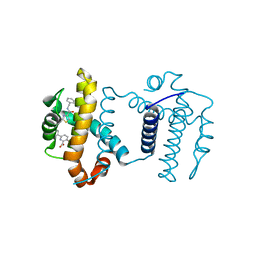 | | Crystal structure of BCL-XL in complex with COMPOUND 1620116, CRYSTAL FORM 1 | | Descriptor: | 6-[(8E)-8-{2-[4-(benzylcarbamoyl)-1,3-thiazol-2-yl]hydrazinylidene}-5,6,7,8-tetrahydronaphthalen-2-yl]-3-(2-phenylethoxy)pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Lee, M, Fairlie, W.D, Smith, B.J, Lee, E.F. | | Deposit date: | 2020-07-19 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
7JGV
 
 | | CRYSTAL STRUCTURE OF BCL-XL IN COMPLEX WITH COMPOUND 1620116, CRYSTAL FORM 2 | | Descriptor: | 6-[(8E)-8-{2-[4-(benzylcarbamoyl)-1,3-thiazol-2-yl]hydrazinylidene}-5,6,7,8-tetrahydronaphthalen-2-yl]-3-(2-phenylethoxy)pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Lee, M, Fairlie, W.D, Smith, B.J, Lee, E.F. | | Deposit date: | 2020-07-19 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
5WPA
 
 | | Structure of human SFPQ/PSPC1 heterodimer | | Descriptor: | Paraspeckle component 1, Splicing factor, proline- and glutamine-rich | | Authors: | Lee, M. | | Deposit date: | 2017-08-04 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of a SFPQ/PSPC1 heterodimer provides insights into preferential heterodimerization of human DBHS family proteins.
J. Biol. Chem., 293, 2018
|
|
4OCA
 
 | | Cryatal structure of ArnB K188A complexted with PLP and UDP-Ara4N | | Descriptor: | (2R,3R,4S,5S)-3,4-dihydroxy-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]tetrahydro-2H-pyr an-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate aminotransferase | | Authors: | Sousa, M.C, Lee, M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Substrate Specificity in ArnB. A Key Enzyme in the Polymyxin Resistance Pathway of Gram-Negative Bacteria.
Biochemistry, 53, 2014
|
|
1FUR
 
 | |
2FUS
 
 | |
