8WRK
 
 | | glycosyltransferase UGT74AN3 | | Descriptor: | Glycosyltransferase, PICEATANNOL, URIDINE-5'-DIPHOSPHATE | | Authors: | Huang, W, Long, F. | | Deposit date: | 2023-10-15 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8H4J
 
 | | Crystal Structure of AzoR-FMN-Lyb24 complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN dependent NADH:quinone oxidoreductase, ~{N}-(3-methylsulfanylphenyl)-4~{H}-cyclopenta[b]quinolin-9-amine | | Authors: | Huang, W. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of AzoR-FMN-Lyb24 complex
To Be Published
|
|
8IND
 
 | | Crystal structure of UGT74AN3-UDP-RES | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, ... | | Authors: | Huang, W. | | Deposit date: | 2023-03-09 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
7DUC
 
 | | Crystal Structure of cyto WalK | | Descriptor: | Sensor protein kinase WalK, ZINC ION | | Authors: | Huang, W. | | Deposit date: | 2021-01-08 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal Structure of cyto WalK
To Be Published
|
|
7DUD
 
 | | Crystal Structure of erWalK | | Descriptor: | Sensor protein kinase WalK | | Authors: | Huang, W. | | Deposit date: | 2021-01-08 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of erWalK
To Be Published
|
|
2M75
 
 | |
2M7H
 
 | |
2M7F
 
 | |
2JTC
 
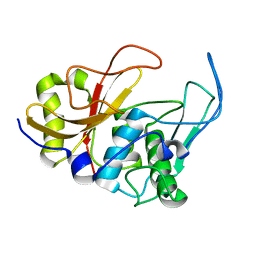 | | 3D structure and backbone dynamics of SPE B | | Descriptor: | Streptopain | | Authors: | Chuang, W, Wang, C, Houng, H, Chen, C, Wang, P. | | Deposit date: | 2007-07-26 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of streptopain: insight into diverse substrate specificity.
J.Biol.Chem., 284, 2009
|
|
8CYA
 
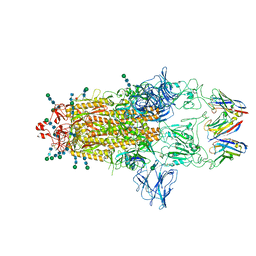 | | SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-67 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Superimmunity by pan-sarbecovirus nanobodies.
Cell Rep, 39, 2022
|
|
8CXQ
 
 | |
8CY6
 
 | |
8CYC
 
 | |
8CYJ
 
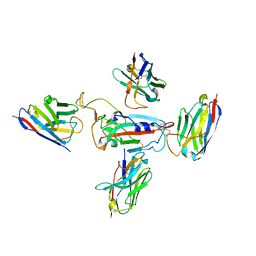 | | RBD of SARS-CoV-2 Spike protein in complex with pan-sarbecovirus nanobodies 2-10, 2-67, 2-62 and 1-25 | | Descriptor: | Spike glycoprotein, pan-sarbecovirus nanobody 1-25, pan-sarbecovirus nanobody 2-10, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Superimmunity by pan-sarbecovirus nanobodies.
Cell Rep, 39, 2022
|
|
8CYB
 
 | |
8CYD
 
 | |
8CY9
 
 | |
8CXN
 
 | |
8CY7
 
 | |
8INV
 
 | | Crystal structure of UGT74AN3-UDP-BUF | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8PFC
 
 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the zinc finger domain of SPL5 from Arabidopsis thaliana | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PFD
 
 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the von Willebrand Factor Type A domain of the proteasomal ubiquitin receptor Rpn10 from Arabidopsis thaliana | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8K5U
 
 | | Se-glycosyltransferase RsSenB | | Descriptor: | Se glycosyltransferase | | Authors: | Huang, W, Long, F. | | Deposit date: | 2023-07-24 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of RsSenB at 2.15 Angstroms resolution
To Be Published
|
|
8IR2
 
 | | Crystal structure of the SLF1 BRCT domain in complex with a Rad18 peptide containing pS442 and pS444 | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Xiang, S, Huang, W, Qiu, F. | | Deposit date: | 2023-03-17 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into Rad18 targeting by the SLF1 BRCT domains.
J.Biol.Chem., 299, 2023
|
|
6UP7
 
 | | neurotensin receptor and arrestin2 complex | | Descriptor: | ARG-ARG-PRO-TYR-ILE-LEU, Beta-arrestin-1, Neurotensin receptor type 1, ... | | Authors: | Qu, Q.H, Huang, W, Masureel, M, Janetzko, J, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2019-10-16 | | Release date: | 2020-02-26 | | Last modified: | 2020-06-17 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the neurotensin receptor 1 in complex with beta-arrestin 1.
Nature, 579, 2020
|
|
