7NAM
 
 | | LRP6_E1 in complex with Lr-EET-3.5 | | Descriptor: | Low-density lipoprotein receptor-related protein 6, SODIUM ION, Trypsin inhibitor 2, ... | | Authors: | Hansen, S, Hannoush, R.N. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directed evolution identifies high-affinity cystine-knot peptide agonists and antagonists of Wnt/ beta-catenin signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XQP
 
 | | PSI-LHCI-LHCII-Lhcb9 supercomplex of Physcomitrella patens | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Zhang, S, Tang, K.L, Li, X.Y, Wang, W.D, Yan, Q.J, Shen, L.L, Kuang, T.Y, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-26 | | Last modified: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural insights into a unique PSI-LHCI-LHCII-Lhcb9 supercomplex from moss Physcomitrium patens.
Nat.Plants, 9, 2023
|
|
5ZCU
 
 | | Crystal structure of RCAR3:PP2C wild-type with pyrabactin | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, Y, Lee, S. | | Deposit date: | 2018-02-20 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural determinants for pyrabactin recognition in ABA receptors in Oryza sativa.
Plant Mol.Biol., 100, 2019
|
|
2QLU
 
 | |
1HAN
 
 | |
1G24
 
 | |
2H2A
 
 | |
2H29
 
 | |
5GWP
 
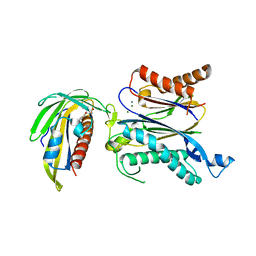 | | Crystal structure of RCAR3:PP2C wild-type with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.577 Å) | | Cite: | Modulation of ABA Signaling by Altering VxG Phi L Motif of PP2Cs in Oryza sativa.
Mol Plant, 10, 2017
|
|
5GWO
 
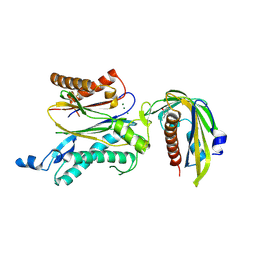 | | Crystal structure of RCAR3:PP2C S265F/I267M with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.816 Å) | | Cite: | Modulation of ABA Signaling by Altering VxG Phi L Motif of PP2Cs in Oryza sativa.
Mol Plant, 10, 2017
|
|
3UE1
 
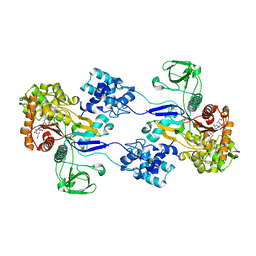 | | Crystal strucuture of Acinetobacter baumanni PBP1A in complex with MC-1 | | Descriptor: | (4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-1-[({4-[(2R)-2,3-dihydroxypropyl]-3-(4,5-dihydroxypyridin-2-yl)-5-oxo-4,5-dihydro- 1H-1,2,4-triazol-1-yl}sulfonyl)amino]-4-formyl-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oate, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UE3
 
 | | Crystal structure of Acinetobacter baumanni PBP3 | | Descriptor: | Septum formation, penicillin binding protein 3, peptidoglycan synthetase | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UDI
 
 | |
3UDF
 
 | | Crystal structure of Apo PBP1a from Acinetobacter baumannii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UE0
 
 | | Crystal structure of Acinetobacter baumannii PBP1a in complex with Aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UDX
 
 | | Crystal structure of Acinetobacter baumannii PBP1a in complex with Imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
4WBC
 
 | | 2.13 A STRUCTURE OF A KUNITZ-TYPE WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN | | Descriptor: | PROTEIN (CHYMOTRYPSIN INHIBITOR), SULFATE ION | | Authors: | Ravichandran, S, Sen, U, Chakrabarti, C, Dattagupta, J.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Cryocrystallography of a Kunitz-type serine protease inhibitor: the 90 K structure of winged bean chymotrypsin inhibitor (WCI) at 2.13 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1T0D
 
 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*(MTU)P*AP*GP*UP*CP*GP*CP*UP*GP*G)-3' | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1T0E
 
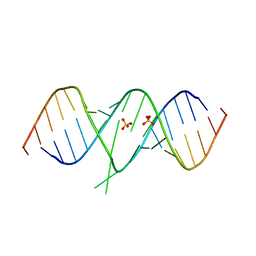 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*GP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*UP*CP*GP*G)-3', SULFATE ION | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
4PJZ
 
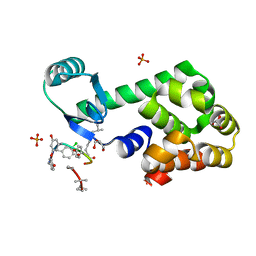 | | CRYSTAL STRUCTURE OF T4 LYSOZYME-GSS-PEPTIDE IN COMPLEX WITH TEICOPLANIN-A2-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, 8-METHYLNONANOIC ACID, ... | | Authors: | Han, S, Le, B.V, Hajare, H, Baxter, R.H.G, Miller, S.J. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray Crystal Structure of Teicoplanin A2-2 Bound to a Catalytic Peptide Sequence via the Carrier Protein Strategy.
J.Org.Chem., 79, 2014
|
|
1QS2
 
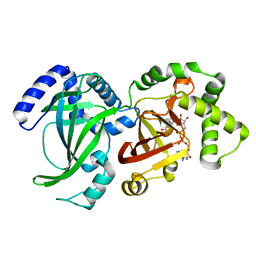 | | CRYSTAL STRUCTURE OF VIP2 WITH NAD | | Descriptor: | ADP-RIBOSYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Han, S, Craig, J.A, Putnam, C.D, Carozzi, N.B, Tainer, J.A. | | Deposit date: | 1999-06-25 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex.
Nat.Struct.Biol., 6, 1999
|
|
1QS1
 
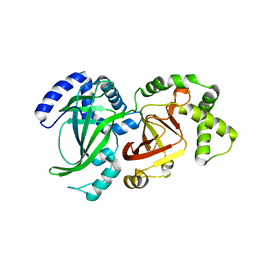 | | CRYSTAL STRUCTURE OF VEGETATIVE INSECTICIDAL PROTEIN2 (VIP2) | | Descriptor: | ADP-RIBOSYLTRANSFERASE | | Authors: | Han, S, Craig, J.A, Putnam, C.D, Carozzi, N.B, Tainer, J.A. | | Deposit date: | 1999-06-25 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex.
Nat.Struct.Biol., 6, 1999
|
|
4PK0
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME-PEPTIDE IN COMPLEX WITH TEICOPLANIN-A2-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, 8-METHYLNONANOIC ACID, ... | | Authors: | Han, S, Le, B.V, Hajare, H, Baxter, R.H.G, Miller, S.J. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure of Teicoplanin A2-2 Bound to a Catalytic Peptide Sequence via the Carrier Protein Strategy.
J.Org.Chem., 79, 2014
|
|
4M14
 
 | |
4M15
 
 | |
