8C2R
 
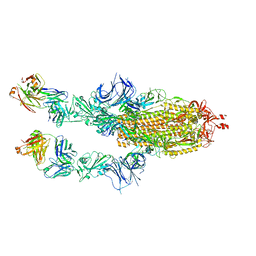 | | SARS-CoV2 Omicron BA.1 spike in complex with CAB-A17 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CAB-A17 antibody (HC), ... | | Authors: | Das, H, Hallberg, B.M. | | Deposit date: | 2022-12-22 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | SARS-CoV-2 omicron Spike (RBD) in complex with CAB-A17 antibody
To Be Published
|
|
4XGM
 
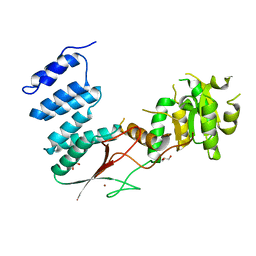 | |
4XGL
 
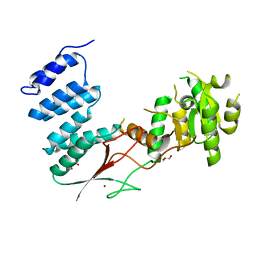 | |
7PZP
 
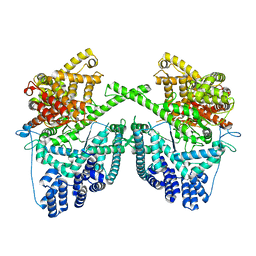 | |
7PZR
 
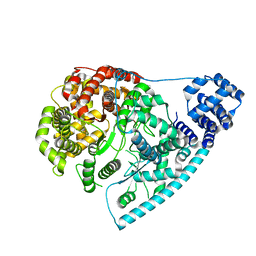 | | Cryo-EM structure of POLRMT in free form. | | Descriptor: | DNA-directed RNA polymerase, mitochondrial | | Authors: | Das, H, Hallberg, B.M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-16 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Non-coding 7S RNA inhibits transcription via mitochondrial RNA polymerase dimerization.
Cell, 185, 2022
|
|
7ZC4
 
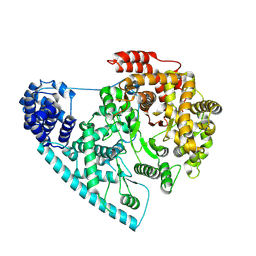 | | Cryo-EM structure of POLRMT mutant. | | Descriptor: | DNA-directed RNA polymerase, mitochondrial | | Authors: | Das, H, Hallberg, B.M. | | Deposit date: | 2022-03-25 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Non-coding 7S RNA inhibits transcription via mitochondrial RNA polymerase dimerization.
Cell, 185, 2022
|
|
8OJC
 
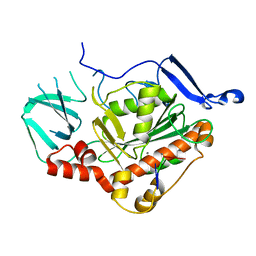 | | HSV-1 DNA polymerase active site in alternative exonuclease state | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA polymerase catalytic subunit | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 2024
|
|
8OJD
 
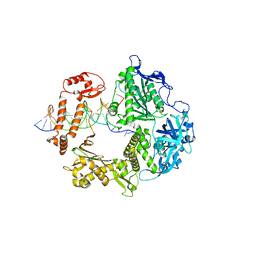 | | HSV-1 DNA polymerase beta-hairpin loop | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA (68-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 2024
|
|
8OJA
 
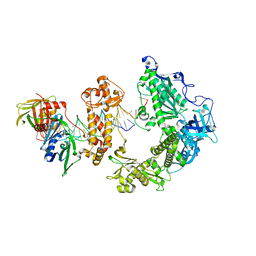 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA (68-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.87 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 2024
|
|
8OJ6
 
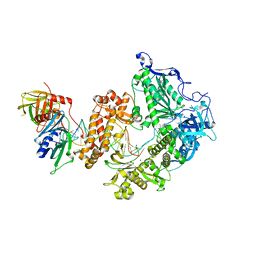 | | HSV-1 DNA polymerase-processivity factor complex in pre-translocation state | | Descriptor: | DNA (22-MER), DNA (48-MER), DNA polymerase catalytic subunit, ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 2024
|
|
8OJ7
 
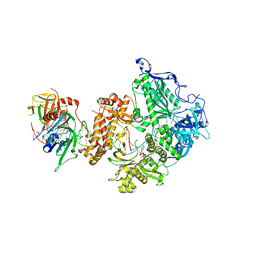 | | HSV-1 DNA polymerase-processivity factor complex in halted elongation state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (53-MER), DNA (67-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 2024
|
|
8OJB
 
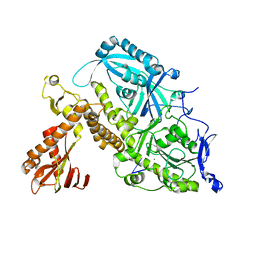 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state active site | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA polymerase catalytic subunit | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 2024
|
|
8P5M
 
 | |
8AS5
 
 | |
3BHD
 
 | | Crystal structure of human thiamine triphosphatase (THTPA) | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Busam, R.D, Lehtio, L, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Weigelt, J, Welin, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Thiamine Triphosphatase.
To be Published
|
|
9ENP
 
 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state with 1-bp DNA mismatch | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (67-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 2024
|
|
9ENQ
 
 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state active site with 1-bp DNA mismatch | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (67-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 2024
|
|
1N8U
 
 | | Chemosensory Protein in Complex with bromo-dodecanol | | Descriptor: | BROMO-DODECANOL, chemosensory protein | | Authors: | Campanacci, V, Lartigue, A, Hallberg, B.M, Jones, T.A, Giudici-Orticoni, M.T, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-21 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1N8V
 
 | | Chemosensory Protein in complex with bromo-dodecanol | | Descriptor: | BROMO-DODECANOL, chemosensory protein | | Authors: | Campanacci, V, Lartigue, A, Hallberg, B.M, Jones, T.A, Giudici-Orticoni, M.T, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-21 | | Release date: | 2003-04-01 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8QVE
 
 | | C-glucosyl oxidoreductase (DaCGO1) from Deinococcus aerius | | Descriptor: | ACETATE ION, CADMIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Furlanetto, V, Kalyani, D.C, Kostelac, A, Haltrich, D, Hallberg, B.M, Divne, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterization of a Gene Cluster Responsible for Deglycosylation of C-glucosyl Flavonoids and Xanthonoids by Deinococcus aerius.
J.Mol.Biol., 436, 2024
|
|
5A2W
 
 | | Crystal structure of mtPAP in complex with ATPgammaS | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, MITOCHONDRIAL PROTEIN, ... | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A30
 
 | | Crystal structure of mtPAP N472D mutant in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, MITOCHONDRIAL PROTEIN, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2Z
 
 | | Crystal structure of mtPAP in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2Y
 
 | | Crystal structure of mtPAP in complex with UTP | | Descriptor: | MAGNESIUM ION, MITOCHONDRIAL PROTEIN, URIDINE 5'-TRIPHOSPHATE | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2X
 
 | | Crystal structure of mtPAP in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
