6XPA
 
 | |
3BKU
 
 | |
3C8F
 
 | |
3BKF
 
 | |
4NJI
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 6-carboxy-5,6,7,8-tetrahydropterin, and Mg2+ | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4NJH
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet and 6-carboxy-5,6,7,8-tetrahydropterin | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
4XC7
 
 | |
4XC6
 
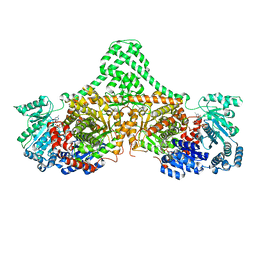 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, and Mg (holo-IcmF/GDP) | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XC8
 
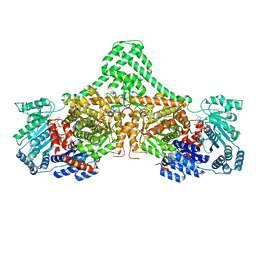 | | Isobutyryl-CoA mutase fused with bound butyryl-CoA, GDP, and Mg and without cobalamin (apo-IcmF/GDP) | | Descriptor: | Butyryl Coenzyme A, GUANOSINE-5'-DIPHOSPHATE, Isobutyryl-CoA mutase fused, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XHM
 
 | |
7KQ4
 
 | |
7KQ3
 
 | |
2R0P
 
 | | K252c-soaked RebC | | Descriptor: | 6,7,12,13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazol-5-one, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QM7
 
 | | MeaB, A Bacterial Homolog of MMAA, Bound to GDP | | Descriptor: | GTPase/ATPase, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Hubbard, P.A, Padovani, D, Labunska, T, Mahlstedt, S.A, Banerjee, R, Drennan, C.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and mutagenesis of the metallochaperone MeaB: insight into the causes of methylmalonic aciduria.
J.Biol.Chem., 282, 2007
|
|
2R0C
 
 | |
7SXM
 
 | |
7TSJ
 
 | |
6XO3
 
 | | ScoE with alpha-ketoglutarate in an off-site | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Jonnalagadda, R, Drennan, C.L. | | Deposit date: | 2020-07-06 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and crystallographic investigations into isonitrile formation by a nonheme iron-dependent oxidase/decarboxylase.
J.Biol.Chem., 296, 2021
|
|
6XOJ
 
 | |
3UUS
 
 | |
5KDP
 
 | | E491A mutant of choline TMA-lyase | | Descriptor: | Choline trimethylamine-lyase, MALONATE ION, SODIUM ION | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
5L2R
 
 | | Crystal structure of fumarate hydratase from Leishmania major | | Descriptor: | (2S)-2-hydroxybutanedioic acid, DI(HYDROXYETHYL)ETHER, Fumarate hydratase, ... | | Authors: | Feliciano, P.R, Drennan, C.L, Nonato, M.C. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal structure of an Fe-S cluster-containing fumarate hydratase enzyme from Leishmania major reveals a unique protein fold.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3OD2
 
 | | E. coli NikR soaked with excess nickel ions | | Descriptor: | 3-CYCLOHEXYLPROPYL 4-O-ALPHA-D-GLUCOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, NICKEL (II) ION, Nickel-responsive regulatory protein | | Authors: | Phillips, C.M, Schreiter, E.R, Drennan, C.L. | | Deposit date: | 2010-08-10 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of low-affinity nickel binding to the nickel-responsive transcription factor NikR from Escherichia coli.
Biochemistry, 49, 2010
|
|
1T3I
 
 | | Structure of slr0077/SufS, the Essential Cysteine Desulfurase from Synechocystis PCC 6803 | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, Probable cysteine desulfurase, ... | | Authors: | Tirupati, B, Vey, J.L, Drennan, C.L, Bollinger Jr, J.M. | | Deposit date: | 2004-04-26 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of Slr0077/SufS, the essential cysteine desulfurase from Synechocystis sp. PCC 6803.
Biochemistry, 43, 2004
|
|
1R30
 
 | | The Crystal Structure of Biotin Synthase, an S-Adenosylmethionine-Dependent Radical Enzyme | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, Biotin synthase, ... | | Authors: | Berkovitch, F, Nicolet, Y, Wan, J.T, Jarrett, J.T, Drennan, C.L. | | Deposit date: | 2003-09-30 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme.
Science, 303, 2004
|
|
