6LBG
 
 | | Structure of OR51B2 bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Olfactory receptor 51B2, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LE6
 
 | | Structure of LNLPTQGRAR bound FEM1C | | Descriptor: | Protein fem-1 homolog C,10-mer peptide, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-24 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LBF
 
 | | Crystal structure of FEM1B | | Descriptor: | Protein fem-1 homolog B, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.252 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LDP
 
 | | Structure of CDK5R1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Cyclin-dependent kinase 5 activator 1, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-22 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LEN
 
 | | Structure of NS11 bound FEM1C | | Descriptor: | Protein fem-1 homolog C,NS11 peptide | | Authors: | Chen, x, Liao, S, Xu, C. | | Deposit date: | 2019-11-25 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
4QDJ
 
 | |
4QDK
 
 | |
2OZ4
 
 | | Structural Plasticity in IgSF Domain 4 of ICAM-1 Mediates Cell Surface Dimerization | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB FRAGMENT LIGHT CHAIN, ... | | Authors: | Chen, X, Kim, T.D, Carman, C.V, Mi, L, Song, G, Springer, T.A. | | Deposit date: | 2007-02-23 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity in Ig superfamily domain 4 of ICAM-1 mediates cell surface dimerization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6WJN
 
 | |
2QTO
 
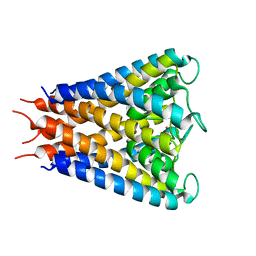 | | An anisotropic model for potassium channel KcsA | | Descriptor: | POTASSIUM ION, Voltage-gated potassium channel | | Authors: | Chen, X, Poon, B.K, Dousis, A, Wang, Q, Ma, J. | | Deposit date: | 2007-08-02 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Normal-mode refinement of anisotropic thermal parameters for potassium channel KcsA at 3.2 A crystallographic resolution
Structure, 15, 2007
|
|
4XKC
 
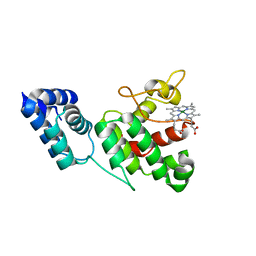 | |
4XKB
 
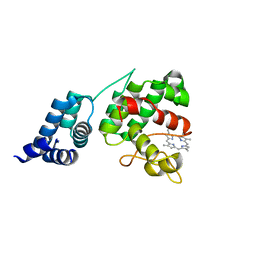 | | Crystal Structure of GENOMES UNCOUPLED 4 (GUN4) in Complex with Deuteroporphyrin IX | | Descriptor: | 3,3'-(3,7,12,17-tetramethylporphyrin-2,18-diyl)dipropanoic acid, Ycf53-like protein | | Authors: | Chen, X, Pu, H, Liu, L. | | Deposit date: | 2015-01-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal Structures of GUN4 in Complex with Porphyrins.
Mol Plant, 8, 2015
|
|
4ZHJ
 
 | |
474D
 
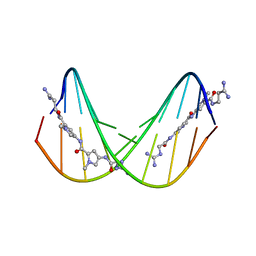 | | A NOVEL END-TO-END BINDING OF TWO NETROPSINS TO THE DNA DECAMER D(CCCCCIIIII)2 | | Descriptor: | DNA (5'-D(*CP*CP*CP*(CBR)P*CP*IP*IP*IP*IP*I)-3'), NETROPSIN | | Authors: | Chen, X, Rao, S.T, Sekar, K, Sundaralingam, M. | | Deposit date: | 1998-01-14 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel end-to-end binding of two netropsins to the DNA decamers d(CCCCCIIIII)2, d(CCCBr5CCIIIII)2and d(CBr5CCCCIIIII)2.
Nucleic Acids Res., 26, 1998
|
|
6OEQ
 
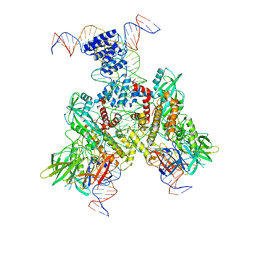 | | Cryo-EM structure of mouse RAG1/2 12RSS-PRC/23RSS-NFC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEO
 
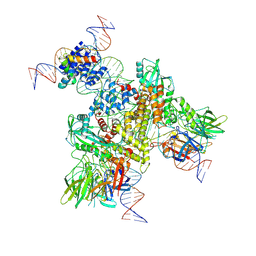 | | Cryo-EM structure of mouse RAG1/2 NFC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OES
 
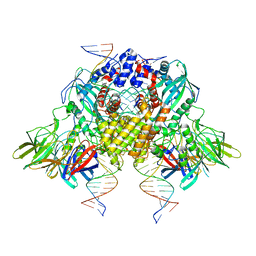 | | Cryo-EM structure of mouse RAG1/2 STC complex (without NBD domain) | | Descriptor: | CALCIUM ION, DNA (34-MER), DNA (35-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | How mouse RAG recombinase avoids DNA transposition.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEP
 
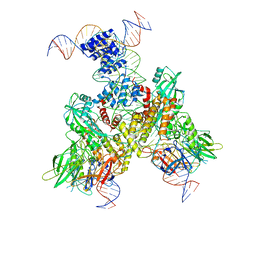 | | Cryo-EM structure of mouse RAG1/2 12RSS-NFC/23RSS-PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OET
 
 | | Cryo-EM structure of mouse RAG1/2 STC complex | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (39-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | How mouse RAG recombinase avoids DNA transposition.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEN
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEM
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA0) | | Descriptor: | DNA (46-MER), DNA (57-MER), High mobility group protein B1, ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OER
 
 | | Cryo-EM structure of mouse RAG1/2 NFC complex (DNA2) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6V0V
 
 | | Cryo-EM structure of mouse WT RAG1/2 NFC complex (DNA0) | | Descriptor: | CALCIUM ION, DNA (30-MER), V(D)J recombination-activating protein 1, ... | | Authors: | Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2019-11-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WJD
 
 | |
6K9A
 
 | | The complex of NrS-1 N terminal domain (1-305) with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Primase | | Authors: | Chen, X, Gan, J. | | Deposit date: | 2019-06-14 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies reveal a unique ring-shaped helicase architecture at the C-terminus of deep-sea vent phage DNA polymerase
To Be Published
|
|
