7KXL
 
 | | BTK1 SOAKED WITH COMPOUND 5, Y551 IS SEQUESTERED | | Descriptor: | 3-tert-butyl-N-({2-fluoro-4-[2-(1-methyl-1H-pyrazol-4-yl)-1H-imidazo[4,5-b]pyridin-7-yl]phenyl}methyl)-1,2,4-oxadiazole-5-carboxamide, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of potent and selective reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 40, 2021
|
|
7KXO
 
 | | BTK1 SOAKED WITH COMPOUND 24 | | Descriptor: | 3-tert-butyl-N-[(1R)-6-{2-[5-methyl-1-(oxan-4-yl)-1H-pyrazol-4-yl]-3H-imidazo[4,5-b]pyridin-7-yl}-1,2,3,4-tetrahydronaphthalen-1-yl]-1,2,4-oxadiazole-5-carboxamide, DIMETHYL SULFOXIDE, Isoform BTK-C of Tyrosine-protein kinase BTK | | Authors: | Viacava Follis, A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of potent and selective reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 40, 2021
|
|
7KXP
 
 | | BTK1 SOAKED WITH COMPOUND 25 | | Descriptor: | 3-tert-butyl-N-[(1S)-6-{2-[3-methyl-1-(oxan-4-yl)-1H-pyrazol-4-yl]-1H-imidazo[4,5-b]pyridin-7-yl}-1,2,3,4-tetrahydronaphthalen-1-yl]-1,2,4-oxadiazole-5-carboxamide, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Viacava Follis, A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of potent and selective reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 40, 2021
|
|
7KXN
 
 | | BTK1 SOAKED WITH COMPOUND 26 | | Descriptor: | 3-tert-butyl-N-[(1S)-6-{2-[5-methyl-1-(propan-2-yl)-1H-pyrazol-4-yl]-1H-imidazo[4,5-b]pyridin-7-yl}-1,2,3,4-tetrahydronaphthalen-1-yl]-1,2,4-oxadiazole-5-carboxamide, DIMETHYL SULFOXIDE, Isoform BTK-C of Tyrosine-protein kinase BTK | | Authors: | Viacava Follis, A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Discovery of potent and selective reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 40, 2021
|
|
7KXM
 
 | | BTK1 SOAKED WITH COMPOUND 5, Y551 IS SEQUESTERED | | Descriptor: | 4-tert-butyl-N-(2-methyl-3-{2-[4-(morpholine-4-carbonyl)phenyl]-1H-imidazo[4,5-b]pyridin-7-yl}phenyl)benzamide, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of potent and selective reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 40, 2021
|
|
7KXQ
 
 | | BTK1 SOAKED WITH COMPOUND 30 | | Descriptor: | 3-tert-butyl-N-[(5R)-2-{2-[3,5-dimethyl-1-(propan-2-yl)-1H-pyrazol-4-yl]-3H-imidazo[4,5-b]pyridin-7-yl}-6,7,8,9-tetrahydro-5H-benzo[7]annulen-5-yl]-1,2,4-oxadiazole-5-carboxamide, DIMETHYL SULFOXIDE, Isoform BTK-C of Tyrosine-protein kinase BTK | | Authors: | Viacava Follis, A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery of potent and selective reversible Bruton's tyrosine kinase inhibitors.
Bioorg.Med.Chem., 40, 2021
|
|
8HGR
 
 | | The apo-flavodoxin monomer from Synechococcus elongatus PCC 7942 | | Descriptor: | CHLORIDE ION, Flavodoxin, MAGNESIUM ION | | Authors: | Liu, S.W, Chen, Y.Y, Gong, Y, Cao, P. | | Deposit date: | 2022-11-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A dimer-monomer transition captured by the crystal structures of cyanobacterial apo flavodoxin.
Biochem.Biophys.Res.Commun., 639, 2022
|
|
8HGQ
 
 | | The apo-flavodoxin dimer from Synechococcus elongatus PCC 7942 | | Descriptor: | Flavodoxin, PHOSPHATE ION | | Authors: | Liu, S.W, Chen, Y.Y, Gong, Y, Cao, P. | | Deposit date: | 2022-11-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A dimer-monomer transition captured by the crystal structures of cyanobacterial apo flavodoxin.
Biochem.Biophys.Res.Commun., 639, 2022
|
|
6A2H
 
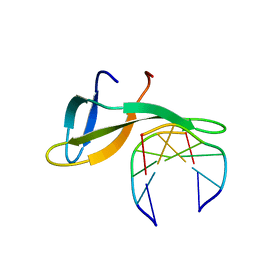 | | Architectural roles of Cren7 in folding crenarchaeal chromatin filament | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(P*AP*AP*TP*TP*AP*C)-3'), DNA (5'-D(P*GP*TP*AP*AP*TP*T)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Chen, Y.Y, Wang, L, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Architectural roles of Cren7 in folding crenarchaeal chromatin filament.
Mol. Microbiol., 111, 2019
|
|
6A2I
 
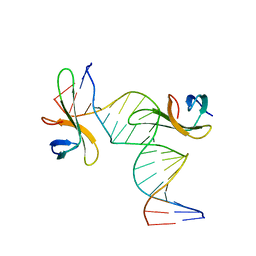 | | Architectural roles of Cren7 in folding crenarchaeal chromatin filament | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*CP*GP*TP*AP*GP*CP*TP*AP*AP*TP*TP*AP*GP*CP*TP*AP*CP*G)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Chen, Y.Y, Wang, L, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architectural roles of Cren7 in folding crenarchaeal chromatin filament.
Mol. Microbiol., 111, 2019
|
|
5YC7
 
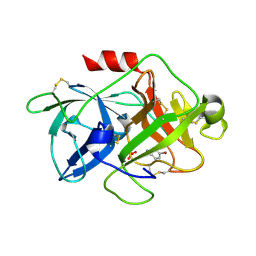 | |
7X8C
 
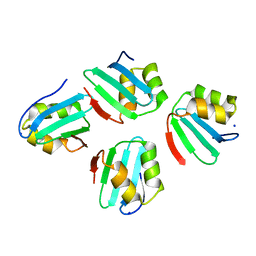 | | Crystal structure of a KTSC family protein from Euryarchaeon Methanolobus vulcani | | Descriptor: | KTSC domain-containing protein, SODIUM ION | | Authors: | Zhang, Z.F, Zhu, K.L, Chen, Y.Y, Cao, P, Gong, Y. | | Deposit date: | 2022-03-12 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Biochemical and structural characterization of a KTSC family single-stranded DNA-binding protein from Euryarchaea.
Int.J.Biol.Macromol., 216, 2022
|
|
5Z1C
 
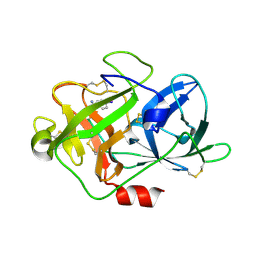 | | The crystal structure of uPA in complex with 4-Iodobenzylamine at pH7.4 | | Descriptor: | 1-(4-iodophenyl)methanamine, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Zhang, X, Luo, Z.P, Huang, M.D. | | Deposit date: | 2017-12-25 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 49, 2018
|
|
3RGU
 
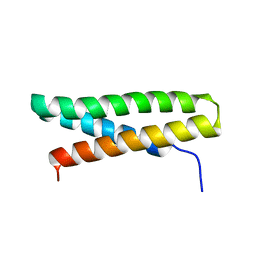 | | Structure of Fap-NRa at pH 5.0 | | Descriptor: | Fimbriae-associated protein Fap1, alpha-D-glucopyranose | | Authors: | Garnett, J.A, Matthews, S.J. | | Deposit date: | 2011-04-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insight into the role of Streptococcus parasanguinis Fap1 within oral biofilm formation.
Biochem.Biophys.Res.Commun., 417, 2012
|
|
5ZLA
 
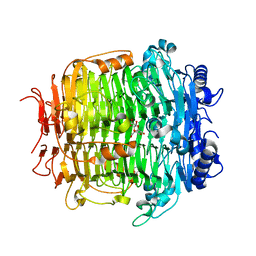 | | Crystal structure of mutant C387A of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-III | | Descriptor: | (2R,3'S,4'S,4aR,5'R,6R,7R,7aS)-4a,5',6-tris(hydroxymethyl)spiro[3,6,7,7a-tetrahydrofuro[2,3-b][1,4]dioxine-2,2'-oxolane ]-3',4',7-triol, DFA-IIIase C387A mutant | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-27 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKW
 
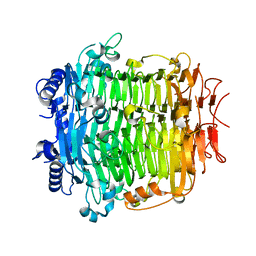 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with GF2 | | Descriptor: | DFA-IIIase, alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZL5
 
 | | Crystal structure of DFA-IIIase mutant C387A from Arthrobacter chlorophenolicus A6 | | Descriptor: | DFA-IIIase C387A mutant, GLYCEROL | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKU
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-III | | Descriptor: | (2R,3'S,4'S,4aR,5'R,6R,7R,7aS)-4a,5',6-tris(hydroxymethyl)spiro[3,6,7,7a-tetrahydrofuro[2,3-b][1,4]dioxine-2,2'-oxolane ]-3',4',7-triol, DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZL4
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 wihout its lid in complex with GF2 | | Descriptor: | DFA-IIIase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKY
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 without its lid | | Descriptor: | DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKS
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 | | Descriptor: | DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
