5IG5
 
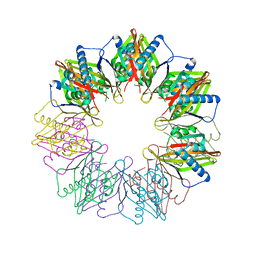 | |
5IG0
 
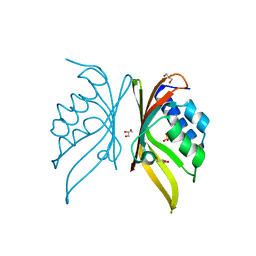 | | Crystal structure of S. rosetta CaMKII hub | | Descriptor: | CAMK/CAMK2 protein kinase, GLYCEROL, SULFATE ION | | Authors: | Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
5IG4
 
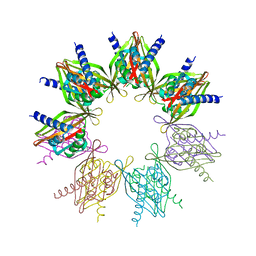 | | Crystal structure of N. vectensis CaMKII-A hub | | Descriptor: | GLYCEROL, Predicted protein | | Authors: | Bhattacharyya, M, Pappireddi, N, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
5IG1
 
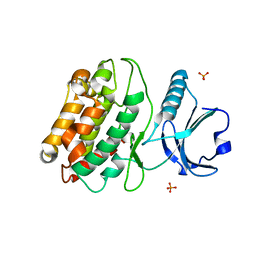 | | Crystal structure of S. rosetta CaMKII kinase domain | | Descriptor: | CAMK/CAMK2 protein kinase, PHOSPHATE ION | | Authors: | Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
6HHC
 
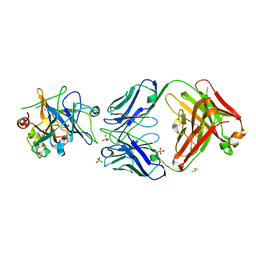 | | Allosteric Inhibition as a new mode of Action for BAY 1213790, a Neutralizing Antibody Targeting the Activated form of Coagulation Factor XI | | Descriptor: | Coagulation factor XI, DIMETHYL SULFOXIDE, FXIA ANTIBODY FAB HEAVY CHAIN, ... | | Authors: | Schaefer, M, Buchmueller, A, Dittmer, F, Strassburger, J, Wilmen, A. | | Deposit date: | 2018-08-27 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric Inhibition as a New Mode of Action for BAY 1213790, a Neutralizing Antibody Targeting the Activated Form of Coagulation Factor XI.
J.Mol.Biol., 431, 2019
|
|
1M1L
 
 | | Human Suppressor of Fused (N-terminal domain) | | Descriptor: | Suppressor of Fused | | Authors: | Merchant, M, Vajdos, F.F, Ultsch, M, Maun, H.R, Wendt, U, Cannon, J, Lazarus, R.A, de Vos, A.M, de Sauvage, F.J. | | Deposit date: | 2002-06-19 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Suppressor of fused regulates Gli activity through a dual binding mechanism
Mol.Cell.Biol., 24, 2004
|
|
4NET
 
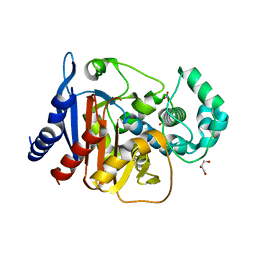 | | Crystal structure of ADC-1 beta-lactamase | | Descriptor: | AmpC, GLYCEROL, NITRATE ION | | Authors: | Bhattacharya, M, Toth, M, Antunes, N.T, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the extended-spectrum class C beta-lactamase ADC-1 from Acinetobacter baumannii.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3O75
 
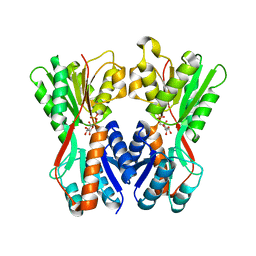 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida in complex with fructose 1-phosphate' | | Descriptor: | 1-O-phosphono-beta-D-fructofuranose, Fructose transport system repressor FruR | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
3O74
 
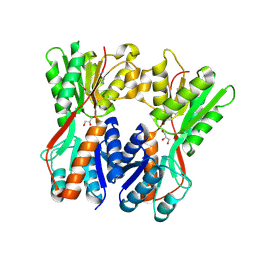 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida | | Descriptor: | Fructose transport system repressor FruR, GLYCEROL | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
3ZN2
 
 | | protein engineering of halohydrin dehalogenase | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETATE ION, HALOHYDRIN DEHALOGENASE, ... | | Authors: | Schallmey, M, Jekel, P, Tang, L, Majeric-Elenkov, M, Hoeffken, H.W, Hauer, B, Janssen, D.B. | | Deposit date: | 2013-02-13 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Single Point Mutation Enhances Hydroxynitrile Synthesis by Halohydrin Dehalogenase.
Enzyme.Microb.Technol., 70, 2015
|
|
1QFQ
 
 | |
1A7Y
 
 | | CRYSTAL STRUCTURE OF ACTINOMYCIN D | | Descriptor: | ACTINOMYCIN D, ETHYL ACETATE, METHANOL | | Authors: | Schafer, M, Sheldrick, G.M, Bahner, I, Lackner, H. | | Deposit date: | 1998-03-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Crystal Structures of Actinomycin D and Actinomycin Z3.
Angew.Chem.Int.Ed.Engl., 37, 1998
|
|
1A7Z
 
 | | CRYSTAL STRUCTURE OF ACTINOMYCIN Z3 | | Descriptor: | ACTINOMYCIN Z3, BENZENE | | Authors: | Schafer, M. | | Deposit date: | 1998-03-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal Structures of Actinomycin D and Actinomycin Z3.
Angew.Chem.Int.Ed.Engl., 37, 1998
|
|
4BGB
 
 | |
4BG9
 
 | | Apo form of a putative sugar kinase MK0840 from Methanopyrus kandleri (orthorhombic space group) | | Descriptor: | ACETATE ION, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Schacherl, M, Waltersperger, S.M, Baumann, U. | | Deposit date: | 2013-03-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Characterization of the Ribonuclease H-Like Type Askha Superfamily Kinase Mk0840 from Methanopyrus Kandleri
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1QGP
 
 | | NMR STRUCTURE OF THE Z-ALPHA DOMAIN OF ADAR1, 15 STRUCTURES | | Descriptor: | PROTEIN (DOUBLE STRANDED RNA ADENOSINE DEAMINASE) | | Authors: | Schade, M, Turner, C.J, Kuehne, R, Schmieder, P, Lowenhaupt, K, Herbert, A, Rich, A, Oschkinat, H. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2BHH
 
 | | HUMAN CYCLIN DEPENDENT PROTEIN KINASE 2 IN COMPLEX WITH THE INHIBITOR 4-HYDROXYPIPERINDINESULFONYL-INDIRUBINE | | Descriptor: | (2E,3S)-3-HYDROXY-5'-[(4-HYDROXYPIPERIDIN-1-YL)SULFONYL]-3-METHYL-1,3-DIHYDRO-2,3'-BIINDOL-2'(1'H)-ONE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Schaefer, M, Jautelat, R, Brumby, T, Briem, H, Eisenbrand, G, Schwahn, S, Krueger, M, Luecking, U, Prien, O, Siemeister, G. | | Deposit date: | 2005-01-11 | | Release date: | 2005-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From the Insoluble Dye Indirubin Towards Highly Active, Soluble Cdk2-Inhibitors
Chembiochem, 6, 2005
|
|
4BGA
 
 | |
2BHE
 
 | | HUMAN CYCLIN DEPENDENT PROTEIN KINASE 2 IN COMPLEX WITH THE INHIBITOR 5-BROMO-INDIRUBINE | | Descriptor: | (2Z)-5'-BROMO-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE AMMONIATE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Schaefer, M, Jautelat, R, Brumby, T, Briem, H, Eisenbrand, G, Schwahn, S, Krueger, M, Luecking, U, Prien, O, Siemeister, G. | | Deposit date: | 2005-01-10 | | Release date: | 2005-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Insoluble Dye Indirubin Towards Highly Active, Soluble Cdk2-Inhibitors
Chembiochem, 6, 2005
|
|
4BG8
 
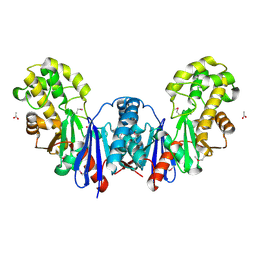 | | Apo form of a putative sugar kinase MK0840 from Methanopyrus kandleri (monoclinic space group) | | Descriptor: | ACETATE ION, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Schacherl, M, Waltersperger, S.M, Baumann, U. | | Deposit date: | 2013-03-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Characterization of the Ribonuclease H-Like Type Askha Superfamily Kinase Mk0840 from Methanopyrus Kandleri
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2BJG
 
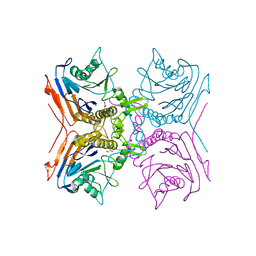 | | Crystal Structure of Conjugated Bile Acid Hydrolase from Clostridium perfringens in Complex with Reaction Products Taurine and Deoxycholate | | Descriptor: | 1,2-ETHANEDIOL, CHOLOYLGLYCINE HYDROLASE | | Authors: | Rossocha, M, Schultz-Heienbrok, R, Von Moeller, H, Coleman, J.P, Saenger, W. | | Deposit date: | 2005-02-02 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conjugated Bile Acid Hydrolase is a Tetrameric N-Terminal Thiol Hydrolase with Specific Recognition of its Cholyl But not of its Tauryl Product
Biochemistry, 44, 2005
|
|
2BJF
 
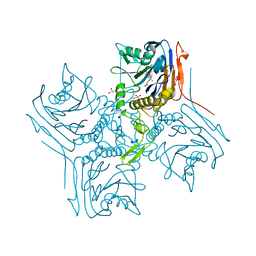 | | Crystal Structure of Conjugated Bile Acid Hydrolase from Clostridium perfringens in Complex with Reaction Products Taurine and Deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, 2-AMINOETHANESULFONIC ACID, CHOLOYLGLYCINE HYDROLASE, ... | | Authors: | Rossocha, M, Schultz-Heienbrok, R, Von Moeller, H, Coleman, J.P, Saenger, W. | | Deposit date: | 2005-02-02 | | Release date: | 2005-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conjugated Bile Acid Hydrolase is a Tetrameric N-Terminal Thiol Hydrolase with Specific Recognition of its Cholyl But not of its Tauryl Product
Biochemistry, 44, 2005
|
|
2LRE
 
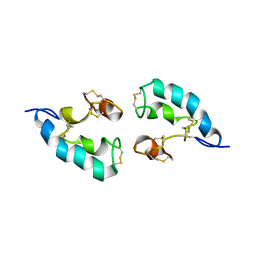 | | The solution structure of the dimeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
2LRD
 
 | | The solution structure of the monomeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
2LD0
 
 | |
