4V6M
 
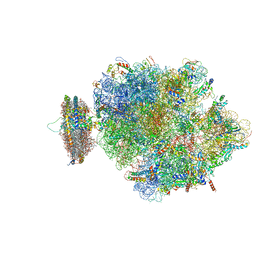 | | Structure of the ribosome-SecYE complex in the membrane environment | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 16S RIBOSOMAL RNA, ... | | Authors: | Frauenfeld, J, Gumbart, J, van der Sluis, E.O, Funes, S, Gartmann, M, Beatrix, B, Mielke, T, Berninghausen, O, Becker, T, Schulten, K, Beckmann, R. | | Deposit date: | 2011-02-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Cryo-EM structure of the ribosome-SecYE complex in the membrane environment.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4UE4
 
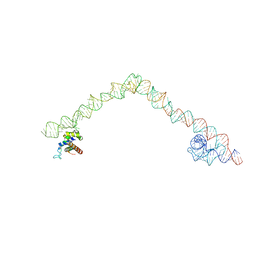 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | Descriptor: | 6S RNA, FTSQ SIGNAL SEQUENCE, SIGNAL RECOGNITION PARTICLE PROTEIN | | Authors: | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UE5
 
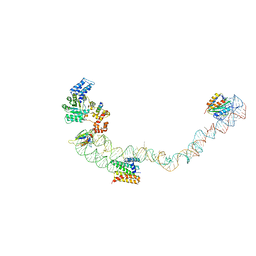 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | Descriptor: | 7S RNA, SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | Authors: | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5A6U
 
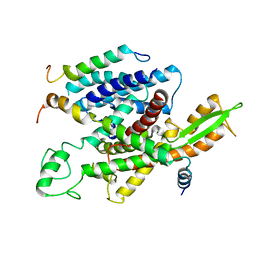 | | Native mammalian ribosome-bound Sec61 protein-conducting channel in the 'non-inserting' state | | Descriptor: | SEC61A, SEC61B, SEC61G | | Authors: | Pfeffer, S, Burbaum, L, Unverdorben, P, Pech, M, Chen, Y, Zimmermann, R, Beckmann, R, Foerster, F. | | Deposit date: | 2015-07-01 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the Native Sec61 Protein-Conducting Channel.
Nat.Commun., 6, 2015
|
|
5A7U
 
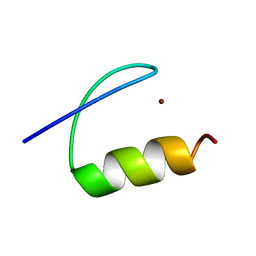 | | Single-particle cryo-EM of co-translational folded adr1 domain inside the E. coli ribosome exit tunnel. | | Descriptor: | REGULATORY PROTEIN ADR1, ZINC ION | | Authors: | Nilsson, O.B, Hedman, R, Marino, J, Wickles, S, Bischoff, L, Johansson, M, Muller-Lucks, A, Trovato, F, Puglisi, J.D, O'Brien, E, Beckmann, R, von Heijne, G. | | Deposit date: | 2015-07-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cotranslational Protein Folding Inside the Ribosome Exit Tunnel.
Cell Rep., 12, 2015
|
|
4V4B
 
 | | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11, ... | | Authors: | Spahn, C.M, Gomez-Lorenzo, M.G, Grassucci, R.A, Jorgensen, R, Andersen, G.R, Beckmann, R, Penczek, P.A, Ballesta, J.P.G, Frank, J. | | Deposit date: | 2004-01-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation.
Embo J., 23, 2004
|
|
4V5H
 
 | | E.Coli 70s Ribosome Stalled During Translation Of Tnac Leader Peptide. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Seidelt, B, Innis, C.A, Wilson, D.N, Gartmann, M, Armache, J, Villa, E, Trabuco, L.G, Becker, T, Mielke, T, Schulten, K, Steitz, T.A, Beckmann, R. | | Deposit date: | 2009-10-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural insight into nascent polypeptide chain-mediated translational stalling.
Science, 326, 2009
|
|
4V6W
 
 | | Structure of the D. melanogaster 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-02-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
4URD
 
 | | Cryo-EM map of Trigger Factor bound to a translating ribosome | | Descriptor: | TRIGGER FACTOR | | Authors: | Deeng, J, Chan, K.Y, van der Sluis, E, Bischoff, L, Berninghausen, O, Han, W, Gumbart, J, Schulten, K, Beatrix, B, Beckmann, R. | | Deposit date: | 2014-06-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Dynamic Behavior of Trigger Factor on the Ribosome.
J.Mol.Biol., 428, 2016
|
|
5ABB
 
 | | Visualization of a polytopic membrane protein during SecY-mediated membrane insertion | | Descriptor: | GREEN-LIGHT ABSORBING PROTEORHODOPSIN, PROTEIN TRANSLOCASE SUBUNIT SECE, PROTEIN TRANSLOCASE SUBUNIT SECY | | Authors: | Bischoff, L, Wickles, S, Berninghausen, O, vanderSluis, E, Beckmann, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Visualization of a Polytopic Membrane Protein During Secy-Mediated Membrane Insertion.
Nat.Commun., 5, 2014
|
|
4V6U
 
 | | Promiscuous behavior of proteins in archaeal ribosomes revealed by cryo-EM: implications for evolution of eukaryotic ribosomes | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10P, ... | | Authors: | Armache, J.-P, Anger, A.M, Marquez, V, Frankenberg, S, Froehlich, T, Villa, E, Berninghausen, O, Thomm, M, Arnold, G.J, Beckmann, R, Wilson, D.N. | | Deposit date: | 2012-08-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Promiscuous behaviour of archaeal ribosomal proteins: Implications for eukaryotic ribosome evolution.
Nucleic Acids Res., 41, 2013
|
|
4V7F
 
 | | Arx1 pre-60S particle. | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Leidig, C, Thoms, M, Holdermann, I, Bradatsch, B, Berninghausen, O, Bange, G, Sinning, I, Hurt, E, Beckmann, R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
4UTQ
 
 | | A structural model of the active ribosome-bound membrane protein insertase YidC | | Descriptor: | ATP SYNTHASE SUBUNIT C, MEMBRANE PROTEIN INSERTASE YIDC | | Authors: | Wickles, S, Singharoy, A, Andreani, J, Seemayer, S, Bischoff, L, Berninghausen, O, Soeding, J, Schulten, K, vanderSluis, E.O, Beckmann, R. | | Deposit date: | 2014-07-22 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A Structural Model of the Active Ribosome-Bound Membrane Protein Insertase Yidc.
Elife, 3, 2014
|
|
4UY8
 
 | | Molecular basis for the ribosome functioning as a L-tryptophan sensor - Cryo-EM structure of a TnaC stalled E.coli ribosome | | Descriptor: | 50S RIBOSOMAL PROTEIN L10, 50S RIBOSOMAL PROTEIN L11, 50S RIBOSOMAL PROTEIN L13, ... | | Authors: | Bischoff, L, Berninghausen, O, Beckmann, R. | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-29 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular Basis for the Ribosome Functioning as an L-Tryptophan Sensor.
Cell Rep., 9, 2014
|
|
6Q8Y
 
 | | Cryo-EM structure of the mRNA translating and degrading yeast 80S ribosome-Xrn1 nuclease complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Heckel, E, Cheng, J, Buschauer, R, Kater, L, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2018-12-16 | | Release date: | 2019-03-13 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the 80S ribosome-Xrn1 nuclease complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8OHD
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ5
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (in-vitro reconstitution) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Peter, J.J, Kulathu, Y, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ8
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ0
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
7OJ0
 
 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide and RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Kudva, R, Becker, T, Berninghausen, O, Heijne, G, Cheng, J, Beckmann, R. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
7OIZ
 
 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Kudva, R, Becker, T, Berninghausen, O, Heijne, G, Cheng, J, Beckmann, R. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
7P3K
 
 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide (control) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Buschauer, R, Komar, T, Becker, T, Berninghausen, O, Cheng, J, Beckmann, R. | | Deposit date: | 2021-07-08 | | Release date: | 2021-10-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
6T4Q
 
 | | Structure of yeast 80S ribosome stalled on the CGA-CCG inhibitory codon combination. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6Z6L
 
 | | Cryo-EM structure of human CCDC124 bound to 80S ribosomes | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
6Z6M
 
 | | Cryo-EM structure of human 80S ribosomes bound to EBP1, eEF2 and SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
