1IMI
 
 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 | | Descriptor: | PROTEIN (ALPHA-CONOTOXIN IMI) | | Authors: | Maslennikov, I.V, Shenkarev, Z.O, Zhmak, M.N, Tsetlin, V.I, Ivanov, V.T, Arseniev, A.S. | | Deposit date: | 1998-11-27 | | Release date: | 1999-04-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR spatial structure of alpha-conotoxin ImI reveals a common scaffold in snail and snake toxins recognizing neuronal nicotinic acetylcholine receptors.
FEBS Lett., 444, 1999
|
|
1NOR
 
 | | TWO-DIMENSIONAL 1H-NMR STUDY OF THE SPATIAL STRUCTURE OF NEUROTOXIN II FROM NAJA OXIANA | | Descriptor: | NEUROTOXIN II | | Authors: | Golovanov, A.P, Utkin, Y.N, Lomize, A.L, Tsetlin, V.I, Arseniev, A.S. | | Deposit date: | 1993-04-05 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Two-dimensional 1H-NMR study of the spatial structure of neurotoxin II from Naja naja oxiana.
Eur.J.Biochem., 213, 1993
|
|
1RQU
 
 | | NMR structure of L7 dimer from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RQS
 
 | | NMR structure of C-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RQT
 
 | | NMR structure of dimeric N-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RQV
 
 | | Spatial model of L7 dimer from E.coli with one hinge region in helical state | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome
J.Biol.Chem., 279, 2004
|
|
1RL5
 
 | | NMR structure with tightly bound water molecule of cytotoxin I from Naja oxiana in aqueous solution (major form) | | Descriptor: | Cytotoxin 1 | | Authors: | Dubinnyi, M.A, Pustovalova, Y.E, Dubovskii, P.V, Utkin, Y.N, Arseniev, A.S. | | Deposit date: | 2003-11-25 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Interaction of three-finger toxins with phospholipid membranes: comparison of S- and P-type cytotoxins.
Biochem.J., 387, 2005
|
|
8AR2
 
 | | Solution structure of TLR5 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 5 | | Authors: | Shabalkina, A.V, Kornilov, F.D, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR1
 
 | | Solution structure of TLR3 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 3 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR3
 
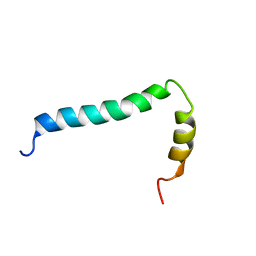 | | Solution structure of TLR9 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 9 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR0
 
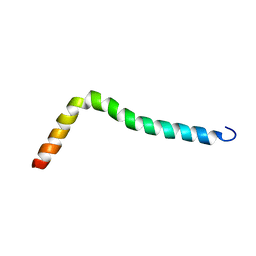 | | Solution structure of TLR2 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 2 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
1BCT
 
 | |
6THI
 
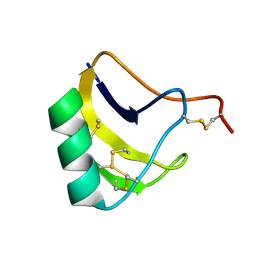 | | Solution structure of MeuNaTxalpha-1 toxin from Mesobuthus Eupeus | | Descriptor: | Sodium channel neurotoxin MeuNaTxalpha-1 | | Authors: | Mineev, K.S, Kuzmenkov, A.I, Khusainov, G.A, Arseniev, A.S, Vassilevski, A.A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of MeuNaTx alpha-1 toxin from scorpion venom highlights the importance of the nest motif.
Proteins, 2021
|
|
5OHD
 
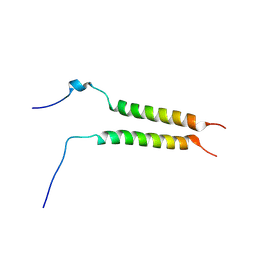 | | Putative inactive (dormant) dimeric state of GHR transmembrane domain | | Descriptor: | Growth hormone receptor | | Authors: | Lesovoy, D.M, Bocharov, E.V, Bocharova, O.V, Urban, A.S, Arseniev, A.S. | | Deposit date: | 2017-07-15 | | Release date: | 2018-04-11 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the signal transduction via transmembrane domain of the human growth hormone receptor.
Biochim. Biophys. Acta, 1862, 2018
|
|
5OEK
 
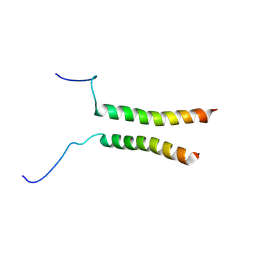 | | Putative active dimeric state of GHR transmembrane domain | | Descriptor: | Growth hormone receptor | | Authors: | Lesovoy, D.M, Bocharov, E.V, Bocharova, O.V, Urban, A.S, Arseniev, A.S. | | Deposit date: | 2017-07-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the signal transduction via transmembrane domain of the human growth hormone receptor.
Biochim. Biophys. Acta, 1862, 2018
|
|
5NQ4
 
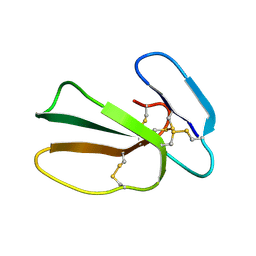 | | Cytotoxin-1 in DPC-micelle | | Descriptor: | Cytotoxin 1 | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Volynsky, P.E, Pustovalova, Y.E, Konshina, A.G, Utkin, Y.N, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-13 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Impact of membrane partitioning on the spatial structure of an S-type cobra cytotoxin.
J. Biomol. Struct. Dyn., 36, 2018
|
|
3MRA
 
 | | M3 TRANSMEMBRANE SEGMENT OF ALPHA-SUBUNIT OF NICOTINIC ACETYLCHOLINE RECEPTOR FROM TORPEDO CALIFORNICA, NMR, 15 STRUCTURES | | Descriptor: | Acetylcholine receptor subunit alpha | | Authors: | Lugovskoy, A.A, Maslennikov, I.V, Utkin, Y.N, Tsetlin, V.I, Cohen, J.B, Arseniev, A.S. | | Deposit date: | 1997-07-15 | | Release date: | 1998-01-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Spatial structure of the M3 transmembrane segment of the nicotinic acetylcholine receptor alpha subunit.
Eur.J.Biochem., 255, 1998
|
|
7QZV
 
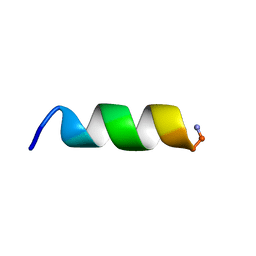 | | Hm-AMP2 | | Descriptor: | Hm-AMP2 | | Authors: | Grafskaia, E.N, Pavlova, E.R, Latsis, I.A, Malakhova, M.V, Lavrenova, V.N, Ivchenkov, D.V, Bashkirov, P.V, Kot, E.F, Mineev, K.S, Arseniev, A.S, Klinov, D.V, Lazarev, V.N. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Non-toxic antimicrobial peptide Hm-AMP2 from leech metagenome proteins identified by the gradient-boosting approach
Materials, 224, 2022
|
|
7QZW
 
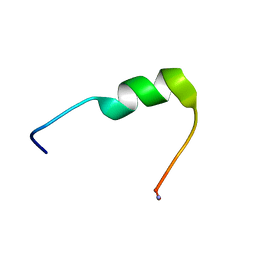 | | Hm-AMP8 | | Descriptor: | Hm-AMP8 | | Authors: | Grafskaia, E.N, Pavlova, E.R, Latsis, I.A, Malakhova, M.V, Lavrenova, V.N, Ivchenkov, D.V, Bashkirov, P.V, Kot, E.F, Mineev, K.S, Arseniev, A.S, Klinov, D.V, Lazarev, V.N. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-15 | | Method: | SOLUTION NMR | | Cite: | The study of the interaction of cationic peptides from a medical leech with blood components as a first step to the systemic use of antimicrobial peptides for therapy of infectious diseases
To Be Published
|
|
6RRO
 
 | |
6RRL
 
 | |
6RSM
 
 | | Solution NMR structure of the peptide 12530 from medicinal leech Hirudo medicinalis in dodecylphosphocholine micelles | | Descriptor: | peptide 12530 | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Grafskaia, E.N, Arseniev, A.S, Lazarev, V.N. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Medicinal leech antimicrobial peptides lacking toxicity represent a promising alternative strategy to combat antibiotic-resistant pathogens.
Eur.J.Med.Chem., 180, 2019
|
|
1BUJ
 
 | |
2N99
 
 | | Solution structure of the SLURP-2, a secreted isoform of Lynx1 | | Descriptor: | Ly-6/neurotoxin-like protein 1 | | Authors: | Paramonov, A.S, Shenkarev, Z.O, Lyukmanova, E.N, Arseniev, A.S. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Secreted Isoform of Human Lynx1 (SLURP-2): Spatial Structure and Pharmacology of Interactions with Different Types of Acetylcholine Receptors.
Sci Rep, 6, 2016
|
|
2N81
 
 | | Solution Structure of Lipid Transfer Protein From Pea Pisum Sativum | | Descriptor: | Lipid Transfer Protein | | Authors: | Paramonov, A.S, Rumynskiy, E.I, Bogdanov, I.V, Finkina, E.I, Melnikova, D.N, Ovchinnikova, T.V, Shenkarev, Z.O, Arseniev, A.S. | | Deposit date: | 2015-09-30 | | Release date: | 2016-05-11 | | Last modified: | 2017-12-20 | | Method: | SOLUTION NMR | | Cite: | A novel lipid transfer protein from the pea Pisum sativum: isolation, recombinant expression, solution structure, antifungal activity, lipid binding, and allergenic properties.
BMC Plant Biol, 16
|
|
