5GOW
 
 | |
3FKR
 
 | | Structure of L-2-keto-3-deoxyarabonate dehydratase complex with pyruvate | | Descriptor: | L-2-keto-3-deoxyarabonate dehydratase, PHOSPHATE ION, SODIUM ION | | Authors: | Shimada, N, Mikami, B. | | Deposit date: | 2008-12-17 | | Release date: | 2010-02-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural analysis of L -2-keto-3-deoxyarabonate dehydratase an enzyme involved in an alternative bacterial pathway of L-arabinose metabolism in complex with pyruvate
To be Published
|
|
7PP2
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-Pii with the host target Exo70F2 from Rice (Oryza sativa) | | Descriptor: | AVR-Pii protein, Exocyst subunit Exo70 family protein, ZINC ION | | Authors: | De la Concepcion, J.C, Bentham, A.R, Lawson, D, Banfield, M.J. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A blast fungus zinc-finger fold effector binds to a hydrophobic pocket in host Exo70 proteins to modulate immune recognition in rice.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3V9Y
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | 4-{4-[({[(9aS)-8-acetyl-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-yl]carbonyl}amino)methyl]naphthalen-2-yl}butanoic acid, Peptide from Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substituents at the naphthalene C3 position of (-)-Cercosporamide derivatives significantly affect the maximal efficacy as PPAR(gamma) partial agonists
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3VVR
 
 | | Crystal structure of MATE in complex with MaD5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein, macrocyclic peptide | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3APQ
 
 | |
3V9T
 
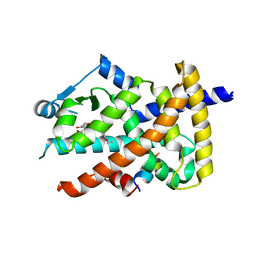 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | (9aS)-8-acetyl-N-[(3-ethoxynaphthalen-1-yl)methyl]-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-carboxamide, Peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Peroxisome proliferator-activated receptor gamma | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substituents at the naphthalene C3 position of (-)-Cercosporamide derivatives significantly affect the maximal efficacy as PPAR(gamma) partial agonists
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3V9V
 
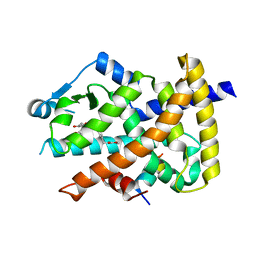 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | Peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Peroxisome proliferator-activated receptor gamma, methyl 3-{4-[({[(9aS)-8-acetyl-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-yl]carbonyl}amino)methyl]naphthalen-2-yl}propanoate | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substituents at the naphthalene C3 position of (-)-Cercosporamide derivatives significantly affect the maximal efficacy as PPAR(gamma) partial agonists
Bioorg.Med.Chem.Lett., 22, 2011
|
|
3APO
 
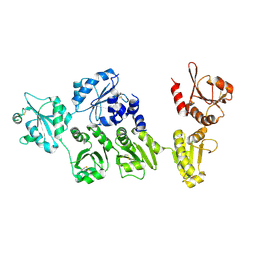 | | Crystal structure of full-length ERdj5 | | Descriptor: | DnaJ homolog subfamily C member 10 | | Authors: | Inaba, K, Suzuki, M, Nagata, K. | | Deposit date: | 2010-10-20 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5.
Mol.Cell, 41, 2011
|
|
3APS
 
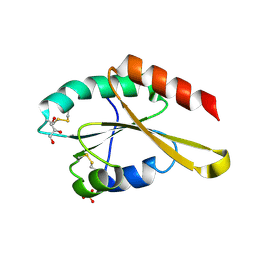 | | Crystal structure of Trx4 domain of ERdj5 | | Descriptor: | DnaJ homolog subfamily C member 10, GLYCEROL, SULFATE ION | | Authors: | Inaba, K, Suzuki, M, Nagata, K. | | Deposit date: | 2010-10-20 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5.
Mol.Cell, 41, 2011
|
|
3VVS
 
 | | Crystal structure of MATE in complex with MaD3S | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein, macrocyclic peptide | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3VVO
 
 | |
3VVN
 
 | |
3A1H
 
 | | Crystal Structure Analysis of the Collagen-like Peptide, (PPG)4-OTG-(PPG)4 | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Miyama, K, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Stabilization of triple-helical structures of collagen peptides containing a Hyp-Thr-Gly, Hyp-Val-Gly, or Hyp-Ser-Gly sequence.
Biopolymers, 95, 2011
|
|
3VVP
 
 | | Crystal structure of MATE in complex with Br-NRF | | Descriptor: | 6-bromo-1-ethyl-4-oxo-7-(piperazin-1-yl)-1,4-dihydroquinoline-3-carboxylic acid, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3WBN
 
 | | Crystal structure of MATE in complex with MaL6 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MaL6, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3WME
 
 | | Crystal structure of an inward-facing eukaryotic ABC multidrug transporter | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6K3D
 
 | | Structure of multicopper oxidase mutant | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, Multicopper oxidase | | Authors: | Sakuraba, H, Ohshida, T, Satomura, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Activity enhancement of multicopper oxidase from a hyperthermophile via directed evolution, and its application as the element of a high performance biocathode.
J.Biotechnol., 325, 2021
|
|
3WUL
 
 | |
3WXS
 
 | | Thaumatin structure determined by SPring-8 Angstrom Compact free electron Laser (SACLA) | | Descriptor: | L(+)-TARTARIC ACID, thaumatin I | | Authors: | Masuda, T, Nango, E, Sugahara, M, Mizohata, E, Tanaka, T, Tanaka, R, Suzuki, M, Mikami, B, Iwata, S. | | Deposit date: | 2014-08-07 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
3WMF
 
 | | Crystal structure of an inward-facing eukaryotic ABC multitrug transporter G277V/A278V/A279V mutant | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WXU
 
 | |
5X2V
 
 | |
5X2Y
 
 | |
5X2W
 
 | |
