3DXI
 
 | |
3DZ1
 
 | |
3DTY
 
 | | Crystal structure of an Oxidoreductase from Pseudomonas syringae | | Descriptor: | MAGNESIUM ION, Oxidoreductase, Gfo/Idh/MocA family | | Authors: | Eswaramoorthy, S, Mahmood, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-16 | | Release date: | 2008-08-05 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of an Oxidoreductase from Pseudomonas syringae
To be Published
|
|
3MSY
 
 | |
3NHM
 
 | |
3NAS
 
 | |
3DZB
 
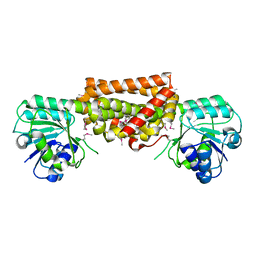 | |
3NIV
 
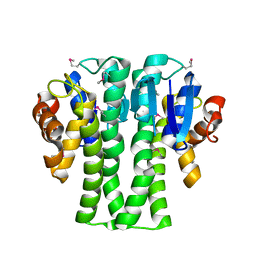 | |
5BUW
 
 | |
2ICS
 
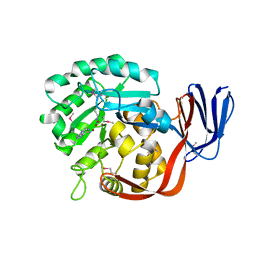 | | Crystal structure of an adenine deaminase | | Descriptor: | ADENINE, Adenine Deaminase, ZINC ION | | Authors: | Sugadev, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an adenine deaminase
TO BE PUBLISHED
|
|
3M2T
 
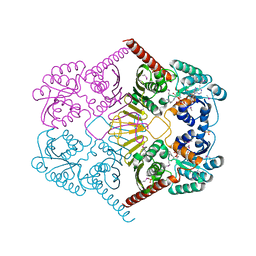 | |
3M3M
 
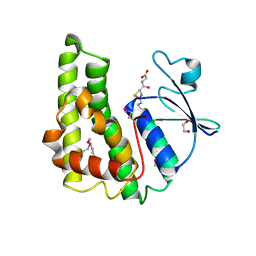 | | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5] | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase, ... | | Authors: | Bagaria, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5]
To be Published
|
|
3FFZ
 
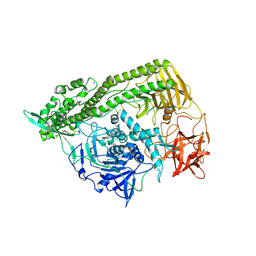 | | Domain organization in Clostridium butulinum neurotoxin type E is unique: Its implication in faster translocation | | Descriptor: | ACETATE ION, Botulinum neurotoxin type E, SODIUM ION, ... | | Authors: | Kumaran, D, Eswaramoorthy, S, Swaminathan, S. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Domain organization in Clostridium botulinum neurotoxin type E is unique: its implication in faster translocation.
J.Mol.Biol., 386, 2009
|
|
2HAF
 
 | | Crystal structure of a putative translation repressor from Vibrio cholerae | | Descriptor: | Putative translation repressor | | Authors: | Sugadev, R, Seetharaman, J, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of a putative translation repressor from Vibrio cholerae
To be Published
|
|
2HAE
 
 | |
3G1W
 
 | |
3EAF
 
 | | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix | | Descriptor: | ABC transporter, substrate binding protein, GLYCEROL, ... | | Authors: | Zhang, Z, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-25 | | Release date: | 2008-09-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix
To be Published
|
|
5BUX
 
 | |
3OR5
 
 | |
3OXN
 
 | |
2I6E
 
 | | Crystal structure of protein DR0370 from Deinococcus radiodurans, Pfam DUF178 | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Tyagi, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of two proteins belonging to Pfam DUF178 revealed unexpected structural similarity to the DUF191 Pfam family.
Bmc Struct.Biol., 7, 2007
|
|
3GBU
 
 | | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Uncharacterized sugar kinase PH1459 | | Authors: | Eswaramoorthy, S, Kumar, G, Zhang, Z, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with ATP
To be Published
|
|
3NQB
 
 | |
3OC4
 
 | | Crystal Structure of a pyridine nucleotide-disulfide family oxidoreductase from the Enterococcus faecalis V583 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase, pyridine nucleotide-disulfide family, ... | | Authors: | Kumaran, D, Baumann, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-09 | | Release date: | 2010-10-06 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a pyridine nucleotide-disulfide family oxidoreductase from the Enterococcus faecalis V583
To be Published
|
|
3G0O
 
 | |
