7VYT
 
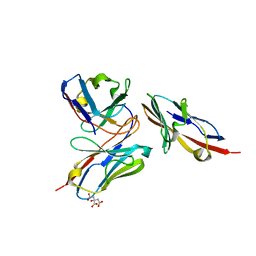 | | Crystal structure of human TIGIT(23-129) in complex with the scFv fragment of anti-TIGIT antibody MG1131 | | Descriptor: | CITRATE ANION, MG1131 heavy chain variable region, MG1131 light chain variable region, ... | | Authors: | Jeong, B.-S, Nam, H, Kim, M, Oh, B.-H. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and functional characterization of a monoclonal antibody blocking TIGIT.
Mabs, 14, 2022
|
|
5X1U
 
 | |
5X1H
 
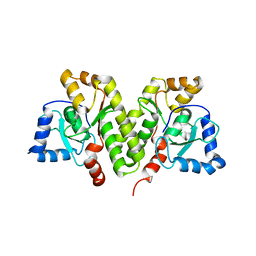 | | Structure of Legionella pneumophila DotN | | Descriptor: | IcmJ (DotN), ZINC ION | | Authors: | Kwak, M.J, Oh, B.H. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
5X1E
 
 | |
3E1W
 
 | |
6KFV
 
 | |
3E1V
 
 | |
5X90
 
 | | Structure of DotL(656-783)-IcmS-IcmW-LvgA derived from Legionella pneumophila | | Descriptor: | Hypothetical virulence protein, IcmO (DotL), IcmS, ... | | Authors: | Kim, H, Kwak, M.J, Kim, J.D, Kim, Y.G, Oh, B.H. | | Deposit date: | 2017-03-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
4J2P
 
 | |
2ESF
 
 | | Identification of a Novel Non-Catalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase | | Descriptor: | BICARBONATE ION, Carbonic anhydrase 2, ZINC ION | | Authors: | Cronk, J.D, Rowlett, R.S, Zhang, K.Y.J, Tu, C, Endrizzi, J.A, Gareiss, P.C. | | Deposit date: | 2005-10-26 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of a Novel Noncatalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase.
Biochemistry, 45, 2006
|
|
5I44
 
 | | Structure of RacA-DNA complex; P21 form | | Descriptor: | Chromosome-anchoring protein RacA, DNA (5'-D(*TP*GP*AP*CP*GP*CP*CP*GP*GP*CP*GP*TP*CP*A)-3') | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Molecular insights into DNA binding and anchoring by the Bacillus subtilis sporulation kinetochore-like RacA protein.
Nucleic Acids Res., 44, 2016
|
|
7JR7
 
 | | Cryo-EM structure of ABCG5/G8 in complex with Fab 2E10 and 11F4 | | Descriptor: | ATP-binding cassette sub-family G member 5, ATP-binding cassette sub-family G member 8, Fab 11F4 heavy chain, ... | | Authors: | Huang, C.S, Yu, X, Min, X, Wang, Z, Zhang, H. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-07 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of ABCG5/G8 in complex with modulating antibodies
Commun Biol, 4, 2021
|
|
4D8H
 
 | | Crystal structure of Symfoil-4P/PV2: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 2 (6xLeu / PV1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo protein | | Authors: | Blaber, M, Longo, L. | | Deposit date: | 2012-01-10 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5I41
 
 | | Structure of the apo RacA DNA binding domain | | Descriptor: | Chromosome-anchoring protein RacA | | Authors: | schumacher, M.A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular insights into DNA binding and anchoring by the Bacillus subtilis sporulation kinetochore-like RacA protein.
Nucleic Acids Res., 44, 2016
|
|
1MRE
 
 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
5K5A
 
 | | Structure of the pNOB8-like ParB N-domain | | Descriptor: | ParB domain protein nuclease | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-15 | | Method: | X-RAY DIFFRACTION (2.825 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5O
 
 | | Structure of AspA-26mer DNA complex | | Descriptor: | AspA, DNA (26-MER) | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5D
 
 | | Structure of the C2221 form of Pnob8-like ParB-N domain | | Descriptor: | CITRIC ACID, ParB domain protein nuclease | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5R
 
 | | AspA-32mer DNA,crystal form 2 | | Descriptor: | AspA, DNA (32-MER), PHOSPHATE ION | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5Q
 
 | |
5IWS
 
 | | Crystal structure of the transporter MalT, the EIIC domain from the maltose-specific phosphotransferase system | | Descriptor: | Protein-N(Pi)-phosphohistidine-sugar phosphotransferase (Enzyme II of the phosphotransferase system) (PTS system glucose-specific IIBC component), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | McCoy, J.G, Ren, Z, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | The Structure of a Sugar Transporter of the Glucose EIIC Superfamily Provides Insight into the Elevator Mechanism of Membrane Transport.
Structure, 24, 2016
|
|
5K5Z
 
 | | Structure of pnob8 ParA | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ParA | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.369 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
4EO6
 
 | |
1MRF
 
 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | 2'-DEOXYINOSINE-5'-MONOPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1MRD
 
 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), IMIDAZOLE, ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
