5YXC
 
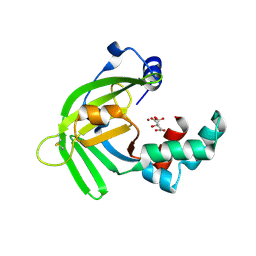 | | Crystal structure of Zinc binding protein ZinT in complex with citrate from E. coli | | Descriptor: | CITRIC ACID, Metal-binding protein ZinT, ZINC ION | | Authors: | Chen, J, Wang, L, Shang, F, Xu, Y. | | Deposit date: | 2017-12-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal structure of E. coli ZinT with one zinc-binding mode and complexed with citrate
Biochem. Biophys. Res. Commun., 500, 2018
|
|
5Z7H
 
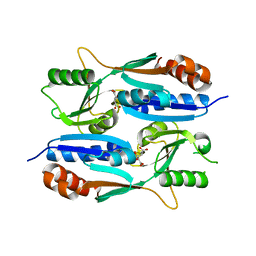 | |
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
2JA1
 
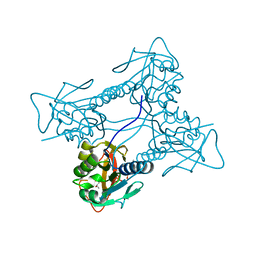 | | Thymidine kinase from B. cereus with TTP bound as phosphate donor. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosinska, U, Carnrot, C, Sandrini, M.P.B, Clausen, A.R, Wang, L, Piskur, J, Eriksson, S, Eklund, H. | | Deposit date: | 2006-11-17 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of Thymidine Kinases from Bacillus Anthracis and Bacillus Cereus Provide Insights Into Quaternary Structure and Conformational Changes Upon Substrate Binding
FEBS J., 274, 2007
|
|
2JAS
 
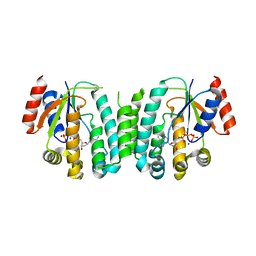 | | Structure of deoxyadenosine kinase from M.mycoides with bound dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DEOXYGUANOSINE KINASE, MAGNESIUM ION | | Authors: | Welin, M, Wang, L, Eriksson, S, Eklund, H. | | Deposit date: | 2006-11-30 | | Release date: | 2007-01-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Function Analysis of a Bacterial Deoxyadenosine Kinase Reveals the Basis for Substrate Specificity.
J.Mol.Biol., 366, 2007
|
|
2J9R
 
 | | Thymidine kinase from B. anthracis in complex with dT. | | Descriptor: | PHOSPHATE ION, THYMIDINE, THYMIDINE KINASE, ... | | Authors: | Kosinska, U, Carnrot, C, Sandrini, M.P.B, Clausen, A.R, Wang, L, Piskur, J, Eriksson, S, Eklund, H. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Studies of Thymidine Kinases from Bacillus Anthracis and Bacillus Cereus Provide Insights Into Quaternary Structure and Conformational Changes Upon Substrate Binding
FEBS J., 274, 2007
|
|
8J3Z
 
 | | Cryo-EM structure of ATP-U46619-bound ABCC4 | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 4, ... | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-04-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8J3W
 
 | | Cryo-EM structure of aspirin-bound ABCC4 | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-04-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
2J9I
 
 | | Lengsin is a survivor of an ancient family of class I glutamine synthetases in eukaryotes that has undergone evolutionary re- engineering for a tissue-specific role in the vertebrate eye lens. | | Descriptor: | GLUTAMATE-AMMONIA LIGASE DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Wyatt, K, White, H.E, Wang, L, Bateman, O.A, Slingsby, C, Orlova, E.V, Wistow, G. | | Deposit date: | 2006-11-09 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Lengsin is a Survivor of an Ancient Family of Class I Glutamine Synthetases Re-Engineered by Evolution for a Role in the Vertebrate Lens.
Structure, 14, 2006
|
|
8IT0
 
 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA dimer (conformation-2) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISZ
 
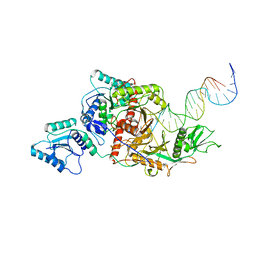 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA monomer | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISY
 
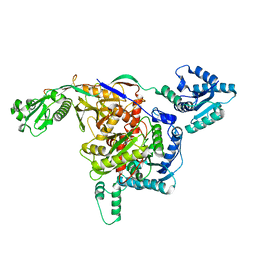 | | Cryo-EM structure of free-state Crt-SPARTA | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8IT1
 
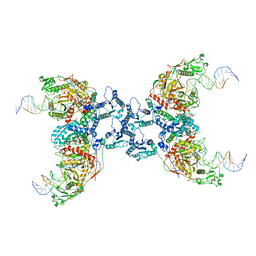 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA tetramer (NADase active form) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8J1K
 
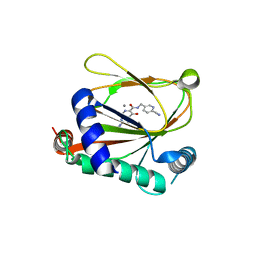 | | co-crystal structure of non-carboxylic acid inhibitor with PHD2 | | Descriptor: | Egl nine homolog 1, MANGANESE (II) ION, N-[(6-cyanopyridin-3-yl)methyl]-5-oxidanyl-2-[(3R)-3-oxidanylpyrrolidin-1-yl]-1,7-naphthyridine-6-carboxamide | | Authors: | Xu, J, Fu, Y, Ding, X, Meng, Q, Wang, L, Zhang, M, Ding, X, Ren, F, Zhavoronkov, A. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | co-crystal structure of non-carboxylic acid inhibitor with PHD2
To Be Published
|
|
8K4Y
 
 | | Structure of a triple-helix region of human ReCol 3 from Trautec | | Descriptor: | SULFATE ION, triple-helix region of human ReCol 3 | | Authors: | Chu, Y, Zhai, Y, Fan, X, Li, J, Wang, L, Fu, S, Feng, P, Qian, S. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a triple-helix region of human ReCol 3 from Trautec.
To Be Published
|
|
8K4X
 
 | | Structure of a triple-helix region of human Collagen type XVII from Trautec | | Descriptor: | triple-helix region of human Collagen type XVII | | Authors: | Chu, Y, Zhai, Y, Fan, X, Li, J, Wang, L, Fu, S, Feng, P, Qian, S. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type XVII from Trautec.
To Be Published
|
|
8K4W
 
 | | Structure of a triple-helix region of human collagen type I from Trautec | | Descriptor: | CHLORIDE ION, triple-helix region of human collagen type I | | Authors: | Chu, Y, Zhai, Y, Fan, X, Li, J, Wang, L, Fu, S, Feng, P, Qian, S. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a triple-helix region of human collagen type I from Trautec.
To Be Published
|
|
8K9G
 
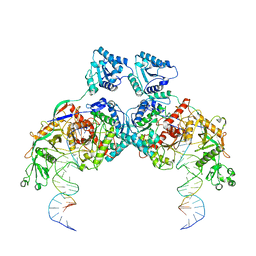 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA dimer (conformation-1) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-08-01 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
2HPL
 
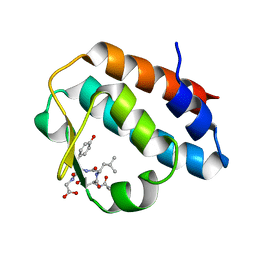 | | Crystal structure of the mouse p97/PNGase complex | | Descriptor: | C-terminal of mouse p97/VCP, PNGase | | Authors: | Zhao, G, Zhou, X, Wang, L, Li, G, Lennarz, W, Schindelin, H. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HPJ
 
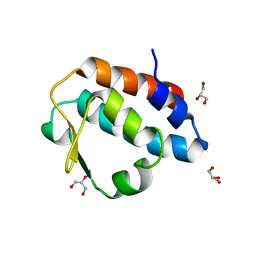 | | Crystal structure of the PUB domain of mouse PNGase | | Descriptor: | GLYCEROL, PNGase | | Authors: | Zhao, G, Zhou, X, Wang, L, Li, G, Lennarz, W, Schindelin, H. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2I74
 
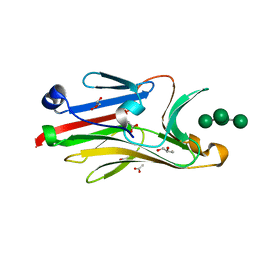 | | Crystal structure of mouse Peptide N-Glycanase C-terminal domain in complex with mannopentaose | | Descriptor: | ACETATE ION, GLYCEROL, PNGase, ... | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2H50
 
 | | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26 | | Descriptor: | small heat shock protein Hsp26 | | Authors: | White, H.E, Orlova, E.V, Chen, S, Wang, L, Ignatiou, A, Gowen, B, Stromer, T, Franzmann, T.M, Haslbeck, M, Buchner, J, Saibil, H.R. | | Deposit date: | 2006-05-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein hsp26
Structure, 14, 2006
|
|
2H53
 
 | | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26 | | Descriptor: | small heat shock protein Hsp26 | | Authors: | White, H.E, Orlova, E.V, Chen, S, Wang, L, Ignatiou, A, Gowen, B, Stromer, T, Franzmann, T.M, Haslbeck, M, Buchner, J, Saibil, H.R. | | Deposit date: | 2006-05-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein hsp26
Structure, 14, 2006
|
|
2JAT
 
 | | Structure of deoxyadenosine kinase from M.mycoides with products dcmp and a flexible dcdp bound | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, DEOXYGUANOSINE KINASE, MAGNESIUM ION, ... | | Authors: | Welin, M, Wang, L, Eriksson, S, Eklund, H. | | Deposit date: | 2006-11-30 | | Release date: | 2007-01-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Function Analysis of a Bacterial Deoxyadenosine Kinase Reveals the Basis for Substrate Specificity.
J.Mol.Biol., 366, 2007
|
|
2JAQ
 
 | | Structure of deoxyadenosine kinase from M. mycoides with bound dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DEOXYGUANOSINE KINASE | | Authors: | Welin, M, Wang, L, Eriksson, S, Eklund, H. | | Deposit date: | 2006-11-30 | | Release date: | 2007-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Function Analysis of a Bacterial Deoxyadenosine Kinase Reveals the Basis for Substrate Specificity.
J.Mol.Biol., 366, 2007
|
|
