5VRE
 
 | |
1MZM
 
 | |
5UV5
 
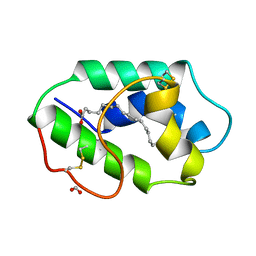
 | | Crystal Structure of a 2-Hydroxyisoquinoline-1,3-dione RNase H Active Site Inhibitor with Multiple Binding Modes to HIV Reverse Transcriptase | | Descriptor: | 7-(furan-2-yl)-2-hydroxyisoquinoline-1,3(2H,4H)-dione, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-02-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A 2-Hydroxyisoquinoline-1,3-Dione Active-Site RNase H Inhibitor Binds in Multiple Modes to HIV-1 Reverse Transcriptase.
Antimicrob. Agents Chemother., 61, 2017
|
|
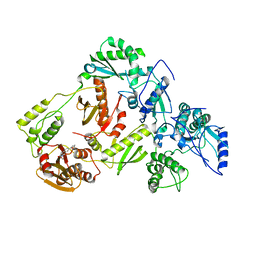
7WAN
 
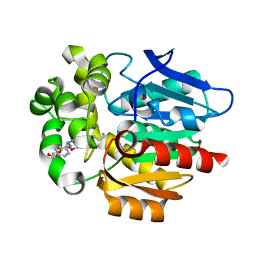 | | Crystal structure of HaloTag complexed with UL2 | | Descriptor: | (R)-[4-(2-azanylhydrazinyl)phenyl]-[2-[2-(2-hexoxyethoxy)ethoxy]ethylamino]methanol, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Pratyush, M, Kang, M, Lee, H, Lee, C, Rhee, H. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.284 Å) | | Cite: | A chemical tool for blue light-inducible proximity photo-crosslinking in live cells.
Chem Sci, 13, 2022
|
|
7WAM
 
 | | Crystal structure of HaloTag complexed with VL1 | | Descriptor: | 3-[6-(2-azanylhydrazinyl)-1,3-bis(oxidanylidene)benzo[de]isoquinolin-2-yl]-N-[2-(2-hexoxyethoxy)ethyl]propanamide, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Pratyush, M, Kang, M, Lee, H, Lee, C, Rhee, H. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A chemical tool for blue light-inducible proximity photo-crosslinking in live cells.
Chem Sci, 13, 2022
|
|
3HM6
 
 | | Crystal structure of the cytoplasmic domain of human plexin B1 | | Descriptor: | Plexin-B1, UNKNOWN ATOM OR ION, Unknown peptide | | Authors: | Tong, Y, He, H, Shen, L, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structure and function of the intracellular region of the plexin-b1 transmembrane receptor.
J.Biol.Chem., 284, 2009
|
|
1XP6
 
 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH COMPOUND 16 | | Descriptor: | (2S,3R)-2-(4-{2-[(3S,4S)-3,4-DIMETHYLPYRROLIDIN-1-YL]ETHOXY}PHENYL)-3-(4-HYDROXYPHENYL)-2,3-DIHYDRO-1,4-BENZOXATHIIN-6-OL, Estrogen receptor | | Authors: | Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2004-10-08 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Estrogen receptor ligands. Part 9: Dihydrobenzoxathiin SERAMs with alkyl substituted pyrrolidine side chains and linkers.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1XP1
 
 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH COMPOUND 15 | | Descriptor: | (2S,3R)-2-(4-{2-[(3R,4R)-3,4-DIMETHYLPYRROLIDIN-1-YL]ETHOXY}PHENYL)-3-(4-HYDROXYPHENYL)-2,3-DIHYDRO-1,4-BENZOXATHIIN-6- OL, Estrogen receptor | | Authors: | Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2004-10-07 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Estrogen receptor ligands. Part 9: Dihydrobenzoxathiin SERAMs with alkyl substituted pyrrolidine side chains and linkers.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1XPC
 
 | |
1XP9
 
 | |
5EEP
 
 | | Crystal structure of E. coli CsdE | | Descriptor: | CsdA-binding activator | | Authors: | Park, S.Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of Escherichia coli CsdE
Int.J.Biol.Macromol., 87, 2016
|
|
5EY0
 
 | | Crystal structure of CodY from Staphylococcus aureus with GTP and Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, GUANOSINE-5'-TRIPHOSPHATE, ISOLEUCINE | | Authors: | Han, A, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
5EY1
 
 | | Crystal structure of CodY from Staphylococcus aureus with GTP and Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, GUANOSINE-5'-TRIPHOSPHATE, ISOLEUCINE | | Authors: | Han, A, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
7EXU
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 E322Q mutant complexed with p-nitrophenyl beta-L-arabinofuranoside | | Descriptor: | (2S,3R,4R,5R)-2-(hydroxymethyl)-5-(4-nitrophenoxy)oxolane-3,4-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Maruyama, S, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXW
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with alpha-L-arabinofuranosylamide | | Descriptor: | 2-bromanyl-N-[(2R,3R,4R,5S}-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXV
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranoylamide | | Descriptor: | 2-bromanyl-N-[(2S,3R,4R,5S)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
2Y2F
 
 | | Crystal structure of Yersinia pestis YopH in complex with an aminooxy- containing platform compound for inhibitor design | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH, [4-[3-(DIFLUORO-PHOSPHONO-METHYL)PHENYL]PHENYL]METHOXYAZANIUM | | Authors: | Lountos, G.T, Bahta, M, Dyas, B, Ulrich, R.G, Waugh, D.S, Burke, T.R. | | Deposit date: | 2010-12-14 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Utilization of Nitrophenylphosphates and Oxime-Based Ligation for the Development of Nanomolar Affinity Inhibitors of the Yersinia Pestis Outer Protein H (Yoph) Phosphatase.
J.Med.Chem., 54, 2011
|
|
4ALR
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALT
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALC
 
 | | X-Ray photoreduction of Polysaccharide monooxigenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALS
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALQ
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALE
 
 | | Structure changes of Polysaccharide monooxygenase CBM33A from Enterococcus faecalis by X-ray induced photoreduction. | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
7DIF
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol at 1.75-angstrom resolution | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, POTASSIUM ION, ... | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3Q6S
 
 | |
