1VHU
 
 | | Crystal structure of a putative phosphoesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hypothetical protein AF1521 | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHZ
 
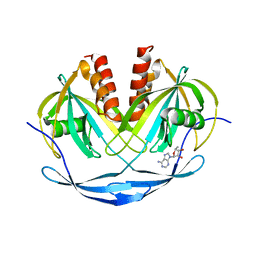 | | Crystal structure of ADP compounds hydrolase | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP compounds hydrolase nudE | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIS
 
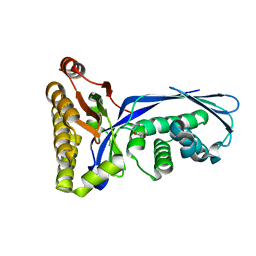 | | Crystal structure of mevalonate kinase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Mevalonate kinase | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VI5
 
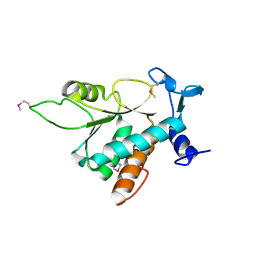 | | Crystal structure of ribosomal protein S2P | | Descriptor: | 30S ribosomal protein S2P | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIZ
 
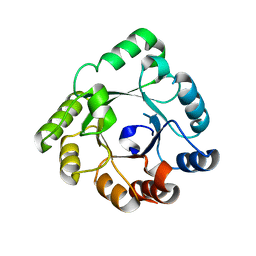 | | Crystal structure of an hypothetical protein | | Descriptor: | PcrB protein homolog, SODIUM ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VH6
 
 | | Crystal structure of a flagellar protein | | Descriptor: | Flagellar protein fliS | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VI9
 
 | | Crystal structure of pyridoxamine kinase | | Descriptor: | BETA-MERCAPTOETHANOL, Pyridoxamine kinase, SULFATE ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHK
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yqeU | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VH2
 
 | |
1VH9
 
 | | Crystal structure of a putative thioesterase | | Descriptor: | Hypothetical protein ybdB | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHG
 
 | | Crystal structure of ADP compounds hydrolase | | Descriptor: | ADP compounds hydrolase nudE | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHT
 
 | |
4GMB
 
 | | Crystal structure of human WD repeat domain 5 with compound MM-402 | | Descriptor: | MM-402, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Cao, F, Liu, L, Lei, M, Dou, Y, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J.Med.Chem., 60, 2017
|
|
5M23
 
 | | Modulation of MLL1 Methyltransferase Activity | | Descriptor: | 4-[(~{E})-[4-[[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-azanyl-3-oxidanyl-propanoyl]amino]propanoyl]amino]-5-carbamimidamido-pentanoyl]amino]propanoyl]amino]methyl]phenyl]diazenyl]-~{N}-[(2~{S})-3-methyl-1-oxidanylidene-butan-2-yl]benzamide, WD repeat-containing protein 5 | | Authors: | Srinivasan, V. | | Deposit date: | 2016-10-11 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Controlled inhibition of methyltransferases using photoswitchable peptidomimetics: towards an epigenetic regulation of leukemia.
Chem Sci, 8, 2017
|
|
6NXF
 
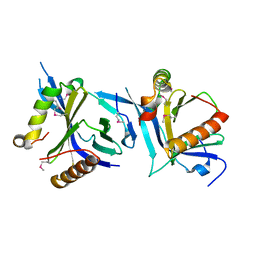 | |
6VYC
 
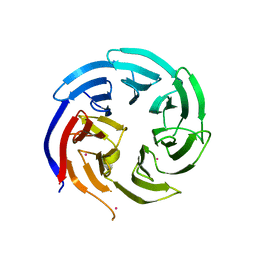 | | Crystal structure of WD-repeat domain of human WDR91 | | Descriptor: | UNKNOWN ATOM OR ION, WD repeat-containing protein 91 | | Authors: | Halabelian, L, Hutchinson, A, Li, Y, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
6C1S
 
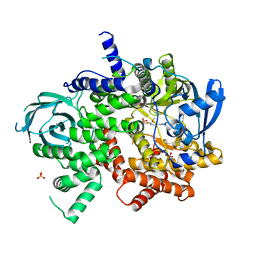 | | Phosphoinositide 3-Kinase gamma bound to an pyrrolopyridinone Inhibitor | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, {4-[2-(5,6-dimethoxypyridin-3-yl)-5-oxo-5,7-dihydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-1H-pyrazol-1-yl}acetonitrile | | Authors: | Jacobs, M.D, Griffin, J.P. | | Deposit date: | 2018-01-05 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design and Synthesis of a Novel Series of Orally Bioavailable, CNS-Penetrant, Isoform Selective Phosphoinositide 3-Kinase gamma (PI3K gamma ) Inhibitors with Potential for the Treatment of Multiple Sclerosis (MS).
J. Med. Chem., 61, 2018
|
|
5CMZ
 
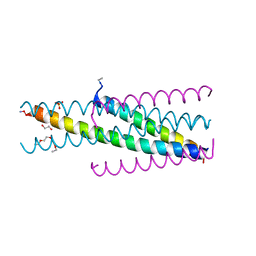 | | Artificial HIV fusion inhibitor AP3 fused to the C-terminus of gp41 NHR | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Artificial HIV entry inhibitor AP3, ... | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CN0
 
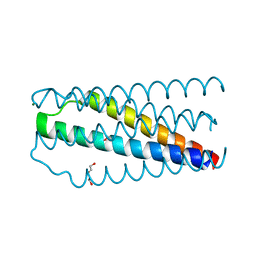 | | Artificial HIV fusion inhibitor AP2 fused to the C-terminus of gp41 NHR | | Descriptor: | DI(HYDROXYETHYL)ETHER, Envelope glycoprotein,AP2, MAGNESIUM ION | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CMU
 
 | |
7U2L
 
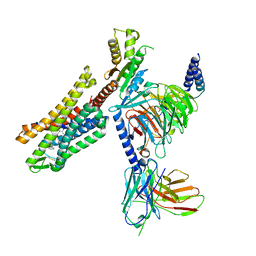 | | C5guano-uOR-Gi-scFv16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Qu, Q, Skiniotis, G, Kobilka, B. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-04 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-based design of bitopic ligands for the μ-opioid receptor.
Nature, 613, 2023
|
|
7U2K
 
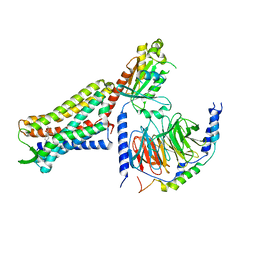 | | C6-guano bound Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Kobilka, B. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based design of bitopic ligands for the μ-opioid receptor.
Nature, 613, 2023
|
|
7KKE
 
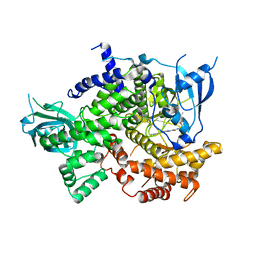 | | Phosphoinositide 3-Kinase gamma bound to a thiazole inhibitor | | Descriptor: | N-[2-(3,3-dimethylbutoxy)ethyl]-N'-{4-methyl-5-[(pyridin-4-yl)ethynyl]-1,3-thiazol-2-yl}urea, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Jacobs, M.D, Griffith, J.P. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Novel Series of Potent and Selective Alkynylthiazole-Derived PI3K gamma Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
4OTF
 
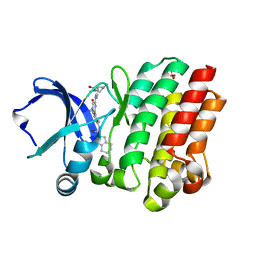 | | Crystal structure of the kinase domain of Bruton's Tyrosine kinase with GDC0834 | | Descriptor: | N-{3-[6-({4-[(2R)-1,4-dimethyl-3-oxopiperazin-2-yl]phenyl}amino)-4-methyl-5-oxo-4,5-dihydropyrazin-2-yl]-2-methylphenyl }-4,5,6,7-tetrahydro-1-benzothiophene-2-carboxamide, SULFATE ION, Tyrosine-protein kinase BTK | | Authors: | Hymowitz, S.G, Maurer, B. | | Deposit date: | 2014-02-13 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent and selective Bruton's tyrosine kinase inhibitors: Discovery of GDC-0834.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4GH6
 
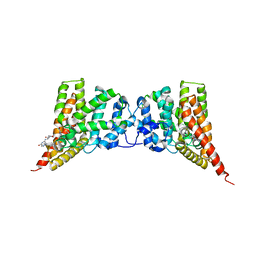 | | Crystal structure of the PDE9A catalytic domain in complex with inhibitor 28 | | Descriptor: | High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, N-(4-methoxyphenyl)-N~2~-[1-(2-methylphenyl)-4-oxo-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl]-L-alaninamide, ... | | Authors: | Hou, J, Ke, H. | | Deposit date: | 2012-08-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Discovery of Highly Selective Phosphodiesterase-9A Inhibitors and Implications for Inhibitor Design.
J.Med.Chem., 55, 2012
|
|
