1MYJ
 
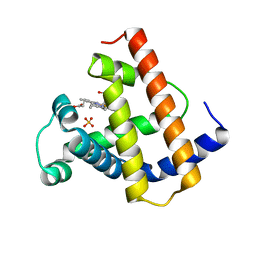 | | DISTAL POLARITY IN LIGAND BINDING TO MYOGLOBIN: STRUCTURAL AND FUNCTIONAL CHARACTERIZATION OF A THREONINE68(E11) MUTANT | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Smerdon, S.J, Oldfield, T.J, Wilkinson, A.J, Dauter, Z, Petratos, K, Wilson, K.S. | | Deposit date: | 1992-02-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distal pocket polarity in ligand binding to myoglobin: structural and functional characterization of a threonine68(E11) mutant.
Biochemistry, 30, 1991
|
|
2JEL
 
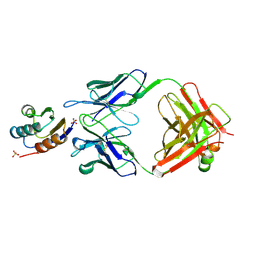 | | JEL42 FAB/HPR COMPLEX | | Descriptor: | HISTIDINE-CONTAINING PROTEIN, JEL42 FAB FRAGMENT, SULFATE ION | | Authors: | Prasad, L, Waygood, E.B, Lee, J.S, Delbaere, L.T.J. | | Deposit date: | 1998-02-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A resolution structure of the jel42 Fab fragment/HPr complex
J.Mol.Biol., 280, 1998
|
|
1P1R
 
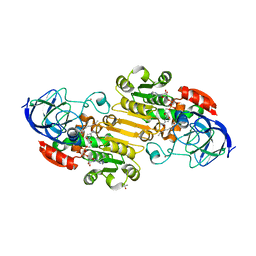 | | Horse liver alcohol dehydrogenase complexed with NADH and R-N-1-methylhexylformamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R)-N-(1-METHYL-HEXYL)-FORMAMIDE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Venkataramaiah, T.H, Plapp, B.V. | | Deposit date: | 2003-04-13 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Formamides mimic aldehydes and inhibit liver alcohol dehydrogenases and ethanol metabolism
J.Biol.Chem., 278, 2003
|
|
1PBF
 
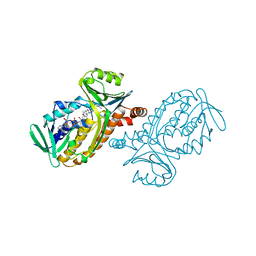 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBD
 
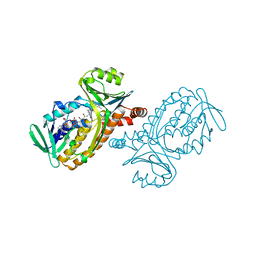 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBB
 
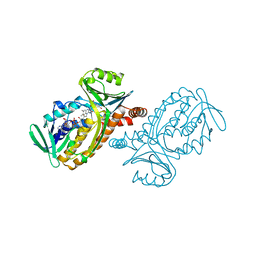 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2,4-DIHYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBC
 
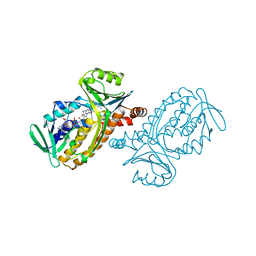 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PDH
 
 | |
2GE5
 
 | |
1NLS
 
 | | CONCANAVALIN A AND ITS BOUND SOLVENT AT 0.94A RESOLUTION | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Deacon, A.M, Gleichmann, T, Helliwell, J.R, Kalb(Gilboa), A.J. | | Deposit date: | 1997-01-28 | | Release date: | 1997-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | The Structure of Concanavalin a and its Bound Solvent Determined with Small-Molecule Accuracy at 0.94 A Resolution
J.Chem.Soc.,Faraday Trans., 93, 1997
|
|
1PZC
 
 | | APO-PSEUDOAZURIN (METAL FREE PROTEIN) | | Descriptor: | PSEUDOAZURIN | | Authors: | Petratos, K. | | Deposit date: | 1995-02-22 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of apo-pseudoazurin from Alcaligenes faecalis S-6.
Febs Lett., 368, 1995
|
|
1SIB
 
 | |
1SBN
 
 | |
2PHH
 
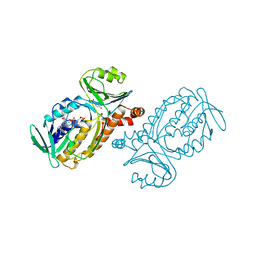 | |
2PPS
 
 | | PHOTOSYNTHETIC REACTION CENTER AND CORE ANTENNA SYSTEM (TRIMERIC), ALPHA CARBON ONLY | | Descriptor: | CHLOROPHYLL A, IRON/SULFUR CLUSTER, PHOTOSYSTEM I, ... | | Authors: | Krauss, N, Schubert, W.-D, Klukas, O, Fromme, P, Witt, H.T, Saenger, W. | | Deposit date: | 1997-05-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Photosystem I at 4 A resolution represents the first structural model of a joint photosynthetic reaction centre and core antenna system.
Nat.Struct.Biol., 3, 1996
|
|
2PSP
 
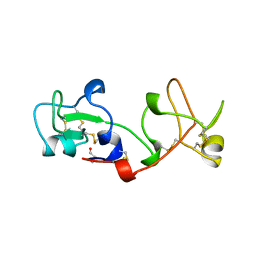 | |
1HSW
 
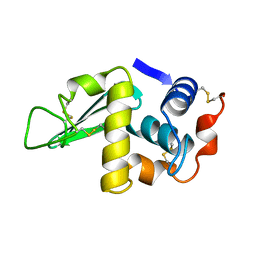 | |
1IGC
 
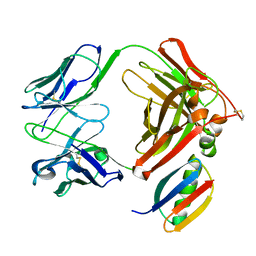 | |
1FFQ
 
 | | CRYSTAL STRUCTURE OF CHITINASE A COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE A | | Authors: | Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo purification scheme and crystallization conditions yield high-resolution structures of chitinase A and its complex with the inhibitor allosamidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1FFR
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT Y390F COMPLEXED WITH HEXA-N-ACETYLCHITOHEXAOSE (NAG)6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1K3H
 
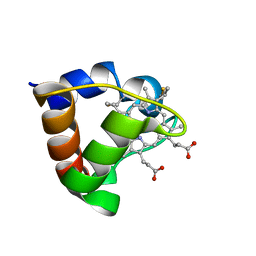 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3G
 
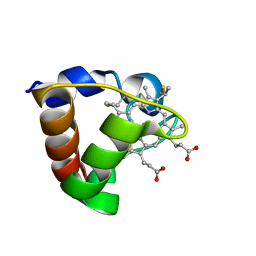 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1GK9
 
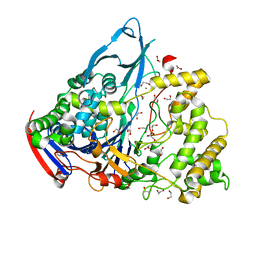 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-08-10 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme-Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1H56
 
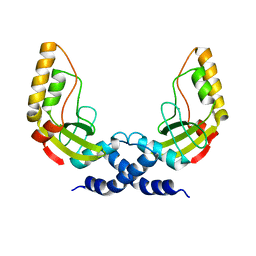 | | Structural and biochemical characterization of a new magnesium ion binding site near Tyr94 in the restriction endonuclease PvuII | | Descriptor: | MAGNESIUM ION, TYPE II RESTRICTION ENZYME PVUII | | Authors: | Spyrida, A, Matzen, C, Lanio, T, Jeltsch, A, Simoncsits, A, Athanasiadis, A, Scheuring-Vanamee, E, Kokkinidis, M, Pingoud, A. | | Deposit date: | 2001-05-20 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Biochemical Characterization of a New Mg(2+) Binding Site Near Tyr94 in the Restriction Endonuclease PvuII.
J.Mol.Biol., 331, 2003
|
|
1H2G
 
 | | Altered substrate specificity mutant of penicillin acylase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Morillas, M, Brannigan, J.A, Ladurner, A.G, Forney, L.J, Virden, R. | | Deposit date: | 2002-08-08 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of Penicillin Acylase Residue B71 Extend Substrate Specificity by Decreasing Steric Constraints for Substrate Binding
Biochem.J., 371, 2003
|
|
