6JJT
 
 | | Crystal structure of an enzyme from Penicillium herquei in condition1 | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
6JJS
 
 | | Crystal structure of an enzyme from Penicillium herquei in condition2 | | Descriptor: | ACETATE ION, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
6KA2
 
 | | Crystal structure of a Thebaine synthase from Papaver somniferum in complex with TBN | | Descriptor: | (4R,7aR,12bS)-7,9-dimethoxy-3-methyl-2,4,7a,13-tetrahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinoline, Thebaine synthase 2 | | Authors: | Xue, J, Yu, X.J, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into thebaine synthase 2 catalysis.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
6KA3
 
 | | Crystal structure of a Thebaine synthase from Papaver somniferum | | Descriptor: | PALMITIC ACID, SULFATE ION, Thebaine synthase 2 | | Authors: | Xue, J, Yu, X.J, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural insights into thebaine synthase 2 catalysis.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
6KQX
 
 | | Crystal structure of Yijc from B. subtilis in complex with UDP | | Descriptor: | URIDINE-5'-DIPHOSPHATE, Uncharacterized UDP-glucosyltransferase YjiC | | Authors: | Hu, Y.M, Dai, L.H, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
6LAA
 
 | | Crystal structure of full-length CYP116B46 from Tepidiphilus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Cytochrome P450, ... | | Authors: | Zhang, L.L, Ko, T.P, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
6KQW
 
 | | Crystal structure of Yijc from B. subtilis | | Descriptor: | CITRIC ACID, Uncharacterized UDP-glucosyltransferase YjiC | | Authors: | Hu, Y.M, Dai, L.H, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
6LDL
 
 | | Crystal structure of CYP116B46-N(20-445) from Tepidiphilus thermophilus in complex with HEME | | Descriptor: | BICINE, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
6LNH
 
 | | Crystal structure of IDO from Bacillus thuringiensis | | Descriptor: | FE (III) ION, L-isoleucine-4-hydroxylase, MERCURY (II) ION | | Authors: | Feng, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of IDO from Bacillus thuringiensis
to be published
|
|
6NCJ
 
 | | Structure of HIV-1 Integrase with potent 5,6,7,8-Tetrahydro-1,6-naphthyridine Derivatives Allosteric Site Inhibitors | | Descriptor: | (2~{S})-2-[4-(8-fluoranyl-5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)-2-methyl-6-[[(1~{S},2~{R})-2-phenylcyclopropyl]methyl]-7,8-dihydro-5~{H}-1,6-naphthyridin-3-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2018-12-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 5,6,7,8-Tetrahydro-1,6-naphthyridine Derivatives as Potent HIV-1-Integrase-Allosteric-Site Inhibitors.
J. Med. Chem., 62, 2019
|
|
2HDD
 
 | | ENGRAILED HOMEODOMAIN Q50K VARIANT DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*GP*GP*GP*GP*AP*TP*TP*AP*CP*AP*TP*GP*G P*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*CP*CP*C P*CP*GP*GP*A)-3'), PROTEIN (ENGRAILED HOMEODOMAIN Q50K) | | Authors: | Tucker-Kellogg, L, Rould, M.A, Chambers, K.A, Ades, S.E, Sauer, R.T, Pabo, C.O. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engrailed (Gln50-->Lys) homeodomain-DNA complex at 1.9 A resolution: structural basis for enhanced affinity and altered specificity.
Structure, 5, 1997
|
|
3CJF
 
 | | Crystal structure of VEGFR2 in complex with a 3,4,5-trimethoxy aniline containing pyrimidine | | Descriptor: | N~4~-(3-methyl-1H-indazol-6-yl)-N~2~-(3,4,5-trimethoxyphenyl)pyrimidine-2,4-diamine, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Nolte, R.T. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of 5-[[4-[(2,3-dimethyl-2H-indazol-6-yl)methylamino]-2-pyrimidinyl]amino]-2-methyl-benzenesulfonamide (Pazopanib), a novel and potent vascular endothelial growth factor receptor inhibitor.
J.Med.Chem., 51, 2008
|
|
3D0X
 
 | |
3CJG
 
 | | Crystal structure of VEGFR2 in complex with a 3,4,5-trimethoxy aniline containing pyrimidine | | Descriptor: | N~4~-methyl-N~4~-(3-methyl-1H-indazol-6-yl)-N~2~-(3,4,5-trimethoxyphenyl)pyrimidine-2,4-diamine, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Nolte, R.T. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of 5-[[4-[(2,3-dimethyl-2H-indazol-6-yl)methylamino]-2-pyrimidinyl]amino]-2-methyl-benzenesulfonamide (Pazopanib), a novel and potent vascular endothelial growth factor receptor inhibitor.
J.Med.Chem., 51, 2008
|
|
3D0U
 
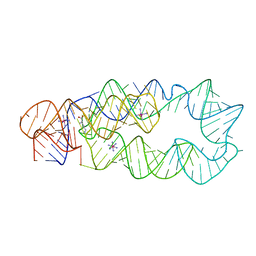 | | Crystal Structure of Lysine Riboswitch Bound to Lysine | | Descriptor: | IRIDIUM HEXAMMINE ION, LYSINE, Lysine Riboswitch RNA | | Authors: | Garst, A.D, Heroux, A, Rambo, R.P, Batey, R.T. | | Deposit date: | 2008-05-02 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the lysine riboswitch regulatory mRNA element.
J.Biol.Chem., 283, 2008
|
|
3DDW
 
 | | Crystal structure of glycogen phosphorylase complexed with an anthranilimide based inhibitor GSK055 | | Descriptor: | (2S)-{[(3-{[(2-chloro-6-methylphenyl)carbamoyl]amino}naphthalen-2-yl)carbonyl]amino}(phenyl)ethanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2008-06-06 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anthranilimide based glycogen phosphorylase inhibitors for the treatment of type 2 diabetes. Part 3: X-ray crystallographic characterization, core and urea optimization and in vivo efficacy.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3DDS
 
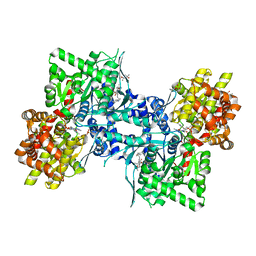 | |
3DD1
 
 | | Crystal structure of glycogen phophorylase complexed with an anthranilimide based inhibitor GSK254 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-cyclohexyl-N-[(3-{[(2,4,6-trimethylphenyl)carbamoyl]amino}naphthalen-2-yl)carbonyl]-D-alanine, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2008-06-04 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Anthranilimide based glycogen phosphorylase inhibitors for the treatment of type 2 diabetes. Part 3: X-ray crystallographic characterization, core and urea optimization and in vivo efficacy.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3DS7
 
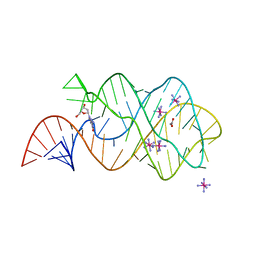 | | Structure of an RNA-2'-deoxyguanosine complex | | Descriptor: | 2'-DEOXY-GUANOSINE, 67-MER, ACETATE ION, ... | | Authors: | Edwards, A.L, Batey, R.T. | | Deposit date: | 2008-07-11 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A structural basis for the recognition of 2'-deoxyguanosine by the purine riboswitch.
J.Mol.Biol., 385, 2009
|
|
3DNJ
 
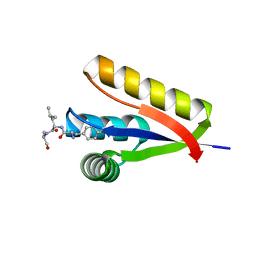 | | The structure of the Caulobacter crescentus ClpS protease adaptor protein in complex with a N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, synthetic N-end rule peptide | | Authors: | Wang, K, Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2008-07-02 | | Release date: | 2008-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The molecular basis of N-end rule recognition.
Mol.Cell, 32, 2008
|
|
3EQ2
 
 | |
3EOD
 
 | |
3ES2
 
 | |
3FO6
 
 | | Crystal structure of guanine riboswitch bound to 6-O-methylguanine | | Descriptor: | 6-O-methylguanine, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Gilbert, S.D, Reyes, F.E, Batey, R.T. | | Deposit date: | 2008-12-28 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adaptive ligand binding by the purine riboswitch in the recognition of Guanine and adenine analogs.
Structure, 17, 2009
|
|
3F7A
 
 | |
