1CLA
 
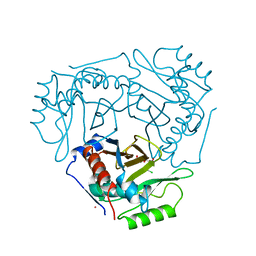 | |
3BFR
 
 | |
3IE5
 
 | | Crystal structure of Hyp-1 protein from Hypericum perforatum (St John's wort) involved in hypericin biosynthesis | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Michalska, K, Fernandes, H, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Crystal structure of Hyp-1, a St. John's wort protein implicated in the biosynthesis of hypericin
J.Struct.Biol., 169, 2010
|
|
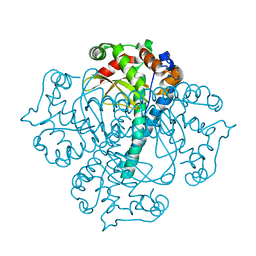
2NO8
 
 | |
2HHN
 
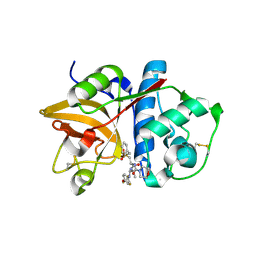 | | Cathepsin S in complex with non covalent arylaminoethyl amide. | | Descriptor: | Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, SULFATE ION | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2006-06-28 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1QMR
 
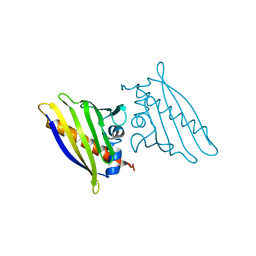 | | BIRCH POLLEN ALLERGEN BET V 1 MUTANT N28T, K32Q, E45S, P108G | | Descriptor: | MAJOR POLLEN ALLERGEN BET V 1-A | | Authors: | Henriksen, A, Holm, J.O, Spangfort, M.D, Gajhede, M. | | Deposit date: | 1999-10-06 | | Release date: | 2000-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Allergy vaccine engineering: epitope modulation of recombinant Bet v 1 reduces IgE binding but retains protein folding pattern for induction of protective blocking-antibody responses.
J Immunol., 173, 2004
|
|
1QFE
 
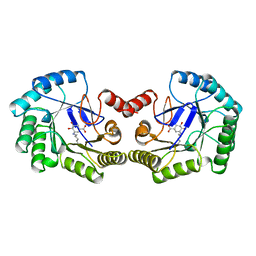 | | THE STRUCTURE OF TYPE I 3-DEHYDROQUINATE DEHYDRATASE FROM SALMONELLA TYPHI | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, PROTEIN (3-DEHYDROQUINATE DEHYDRATASE) | | Authors: | Shrive, A.K, Polikarpov, I, Sawyer, L, Coggins, J.R, Hawkins, A.R. | | Deposit date: | 1999-04-05 | | Release date: | 2000-04-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The two types of 3-dehydroquinase have distinct structures but catalyze the same overall reaction.
Nat.Struct.Biol., 6, 1999
|
|
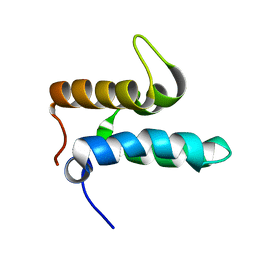
2N3X
 
 | |
1ICX
 
 | |
7FTQ
 
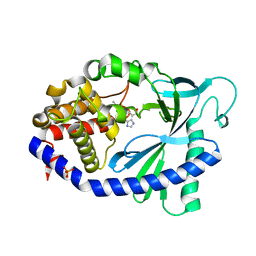 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 6-(2-chloro-5-fluoro-4-methylphenyl)-1H-benzimidazole-4-carboxylic acid | | Descriptor: | (5M)-5-(2-chloro-5-fluoro-4-methylphenyl)-1H-benzimidazole-7-carboxylic acid, Cyclic GMP-AMP synthase, PHOSPHATE ION, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Zhou, C, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
4CLA
 
 | |
7FTO
 
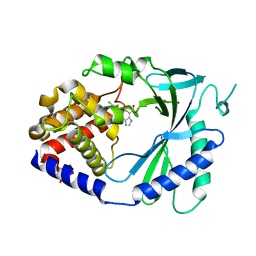 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 6-(2-chloro-5-fluoro-4-methylphenyl)-1H-benzimidazole-4-carboxylic acid | | Descriptor: | (5M)-5-(2-chloro-5-fluoro-4-methylphenyl)-1H-benzimidazole-7-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Zhou, C, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FU2
 
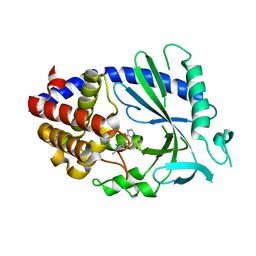 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 6-(2-chloro-5-fluoro-4-methylphenyl)-1H-benzimidazole-4-carboxylic acid | | Descriptor: | (5M)-5-(2-chloro-5-fluoro-4-methylphenyl)-1H-benzimidazole-7-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Zhou, C, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
2CLA
 
 | |
1IFV
 
 | |
2FLH
 
 | | Crystal structure of cytokinin-specific binding protein from mung bean in complex with cytokinin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, SODIUM ION, cytokinin-specific binding protein | | Authors: | Pasternak, O, Bujacz, G.D, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Vigna radiata Cytokinin-Specific Binding Protein in Complex with Zeatin.
Plant Cell, 18, 2006
|
|
