7E1Q
 
 | | Crystal structure of dehydrogenase/isomerase FabX from Helicobacter pylori | | Descriptor: | 2-nitropropane dioxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1R
 
 | | Crystal structure of Dehydrogenase/isomerase FabX from Helicobacter pylori in complex with holo-ACP | | Descriptor: | 2-nitropropane dioxygenase, Acyl carrier protein,Acyl carrier protein, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1S
 
 | | Crystal structure of dehydrogenase/isomerase FabX from Helicobacter pylori in complex with octanoyl-ACP | | Descriptor: | 2-nitropropane dioxygenase, Acyl carrier protein,Acyl carrier protein, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1L
 
 | | Crystal structure of apo form PhlH | | Descriptor: | DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
7E1N
 
 | | Crystal structure of PhlH in complex with 2,4-diacetylphloroglucinol | | Descriptor: | 2,4-bis[(1R)-1-oxidanylethyl]benzene-1,3,5-triol, DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
5FWG
 
 | | TETRA-(5-FLUOROTRYPTOPHANYL)-GLUTATHIONE TRANSFERASE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, TETRA-(5-FLUOROTRYPTOPHANYL)-GLUTATHIONE TRANSFERASE MU CLASS | | Authors: | Parsons, J.F, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1997-11-08 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymes harboring unnatural amino acids: mechanistic and structural analysis of the enhanced catalytic activity of a glutathione transferase containing 5-fluorotryptophan.
Biochemistry, 37, 1998
|
|
5GST
 
 | | REACTION COORDINATE MOTION IN AN SNAR REACTION CATALYZED BY GLUTATHIONE TRANSFERASE | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-07-20 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots along the reaction coordinate of an SNAr reaction catalyzed by glutathione transferase.
Biochemistry, 32, 1993
|
|
8XOM
 
 | | Cryo-EM structure of human ABCC4 in complex with ANP-bound in NBD1 and METHOTREXATE | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, MAGNESIUM ION, ... | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
8XOL
 
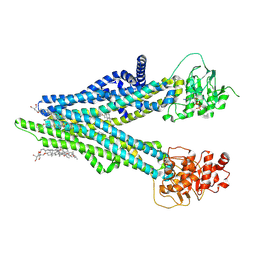 | | Cryo-EM structure of human ABCC4 with ANP bound in NBD1 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, MAGNESIUM ION, ... | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
8XOK
 
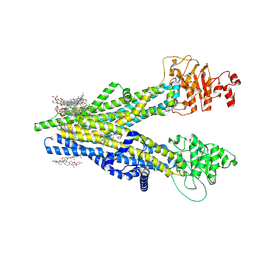 | | Cryo-EM structure of human ABCC4 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, PALMITIC ACID | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
2KOD
 
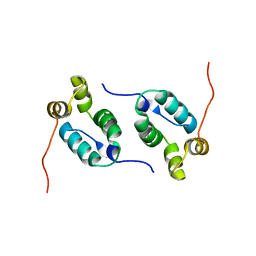 | | A high-resolution NMR structure of the dimeric C-terminal domain of HIV-1 CA | | Descriptor: | HIV-1 CA C-terminal domain | | Authors: | Byeon, I.-J.L, Jung, J, Ahn, J, concel, J, Gronenborn, A.M. | | Deposit date: | 2009-09-18 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function.
Cell(Cambridge,Mass.), 139, 2009
|
|
9CUB
 
 | | Human STING G230A/R293Q variant bound to diABZI-a1 | | Descriptor: | 1,1'-[(2E)-but-2-ene-1,4-diyl]bis{2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-1,3-benzimidazole-5-carboxamide}, Stimulator of interferon genes protein | | Authors: | Critton, D.A. | | Deposit date: | 2024-07-26 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Orthosteric STING inhibition elucidates molecular correction of SAVI STING
To Be Published
|
|
9CUA
 
 | |
9CUD
 
 | | Human STING G230A/R293Q variant bound to diABZI-i | | Descriptor: | (2E)-1-[(2E)-4-{(2E)-5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)imino]-3-methyl-2,3-dihydro-1H-1,3-benzimidazol-1-yl}-2,3-dimethylbut-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)imino]-7-[(3-methoxyphenyl)methoxy]-3-methyl-2,3-dihydro-1H-1,3-benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Critton, D.A. | | Deposit date: | 2024-07-26 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Orthosteric STING inhibition elucidates molecular correction of SAVI STING
To Be Published
|
|
9CUE
 
 | | Human STING H232R variant bound to ABZI | | Descriptor: | Stimulator of interferon genes protein, benzyl (3-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-1,3-benzimidazol-1-yl}propyl)carbamate | | Authors: | Sack, J.S, Critton, D.A. | | Deposit date: | 2024-07-26 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Orthosteric STING inhibition elucidates molecular correction of SAVI STING
To Be Published
|
|
9CUC
 
 | | Human STING G230A/R293Q variant bound to THIQi | | Descriptor: | (3S,4S)-2-(4-tert-butyl-3-chlorophenyl)-3-(2,3-dihydro-1,4-benzodioxin-6-yl)-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxylic acid, Stimulator of interferon genes protein | | Authors: | Critton, D.A. | | Deposit date: | 2024-07-26 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Orthosteric STING inhibition elucidates molecular correction of SAVI STING
To Be Published
|
|
8RLB
 
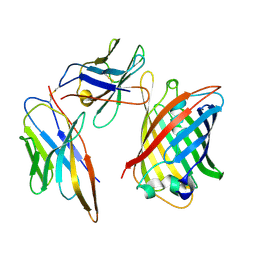 | | RECQL5:sfGFP hetero dimer assembled by Di-Gluebody - sfGFP local refinement | | Descriptor: | Gluebody G5-006, Gluebody GbEnhancer, Green fluorescent protein | | Authors: | Yi, G, Ye, M, Mamalis, D, Fairhead, M, Sauer, D.B, von Delft, F, Davis, B.G, Gilbert, R.J.C. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Di-Gluebodies: Rigid modular nanobody protein assemblies enabling simultaneous determination of high-resolution cryo-EM structures
To Be Published
|
|
8RL8
 
 | | SPNS2 in complex with homo Di-Gluebody GbD12 - SPNS2 local refinement | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Yi, G, Ye, M, Mamalis, D, Sauer, D.B, von Delft, F, Davis, B.G, Gilbert, R.J.C. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Di-Gluebodies: Rigid modular nanobody protein assemblies enabling simultaneous determination of high-resolution cryo-EM structures
To Be Published
|
|
8RLD
 
 | | SPNS2:sfGFP hetero dimer assembled by Di-Gluebody - SPNS2 local refinement | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Yi, G, Ye, M, Mamalis, D, Sauer, D.B, von Delft, F, Davis, B.G, Gilbert, R.J.C. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Di-Gluebodies: Rigid modular nanobody protein assemblies enabling simultaneous determination of high-resolution cryo-EM structures
To Be Published
|
|
8RLC
 
 | | SPNS2:sfGFP hetero dimer assembled by Di-Gluebody | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gluebody GbC4, Gluebody GbEnhancer, ... | | Authors: | Yi, G, Ye, M, Mamalis, D, Li, H, Sauer, D.B, von Delft, F, Davis, B.G, Gilbert, R.J.C. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Di-Gluebodies: Rigid modular nanobody protein assemblies enabling simultaneous determination of high-resolution cryo-EM structures
To Be Published
|
|
8RL7
 
 | | SPNS2 in complex with homo Di-Gluebody GbD12 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gluebody GbD12, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Yi, G, Ye, M, Mamalis, D, Li, H, Sauer, D.B, von Delft, F, Davis, B.G, Gilbert, R.J.C. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Di-Gluebodies: Rigid modular nanobody protein assemblies enabling simultaneous determination of high-resolution cryo-EM structures
To Be Published
|
|
8RL9
 
 | | RECQL5:sfGFP hetero dimer assembled by Di-Gluebody | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, Gluebody GbEnhancer, ... | | Authors: | Yi, G, Ye, M, Mamalis, D, Fairhead, M, Sauer, D.B, von Delft, F, Davis, B.G, Gilbert, R.J.C. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Di-Gluebodies: Rigid modular nanobody protein assemblies enabling simultaneous determination of high-resolution cryo-EM structures
To Be Published
|
|
8RL6
 
 | | DNA helicase RECQL5 in complex with homo Di-Gluebody G5-006 - RECQL5 local refinement | | Descriptor: | ATP-dependent DNA helicase Q5, ZINC ION | | Authors: | Yi, G, Ye, M, Mamalis, D, Sauer, D.B, von Delft, F, Davis, B.G, Gilbert, R.J.C. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Di-Gluebodies: Rigid modular nanobody protein assemblies enabling simultaneous determination of high-resolution cryo-EM structures
To Be Published
|
|
8RLA
 
 | | RECQL5:sfGFP hetero dimer assembled by Di-Gluebody - RECQL5 local refinement | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, ZINC ION | | Authors: | Yi, G, Ye, M, Mamalis, D, Sauer, D.B, von Delft, F, Davis, B.G, Gilbert, R.J.C. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Di-Gluebodies: Rigid modular nanobody protein assemblies enabling simultaneous determination of high-resolution cryo-EM structures
To Be Published
|
|
8RL5
 
 | | DNA helicase RECQL5 in complex with homo Di-Gluebody G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, ZINC ION | | Authors: | Yi, G, Ye, M, Mamalis, D, Sauer, D.B, von Delft, F, Davis, B.G, Gilbert, R.J.C. | | Deposit date: | 2024-01-02 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Di-Gluebodies: Rigid modular nanobody protein assemblies enabling simultaneous determination of high-resolution cryo-EM structures
To Be Published
|
|
