8U15
 
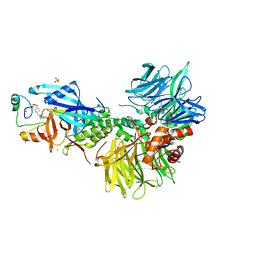 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-short bound to CC-220 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DDB1, Protein cereblon, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
7X0T
 
 | |
7X0Z
 
 | |
7X07
 
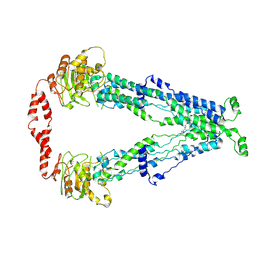 | | Cryo-EM structure of human ABCD1 in the presence of C26:0 | | Descriptor: | ATP-binding cassette sub-family D member 1, hexacosanoic acid | | Authors: | Chao, X, Li-Na, J, Lin, T. | | Deposit date: | 2022-02-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural insights into substrate recognition and translocation of human peroxisomal ABC transporter ALDP.
Signal Transduct Target Ther, 8, 2023
|
|
7X1W
 
 | |
7XEC
 
 | |
6GDO
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 2g (F240) | | Descriptor: | ACETATE ION, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
8IFZ
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2023-02-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8IFY
 
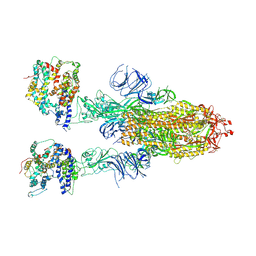 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 spike protein in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2023-02-20 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
6GD0
 
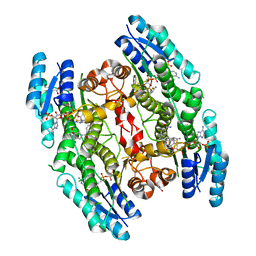 | | Trypanosoma brucei PTR1 in complex with inhibitor 4g (F133) | | Descriptor: | ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase, ... | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2018-04-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
6GCK
 
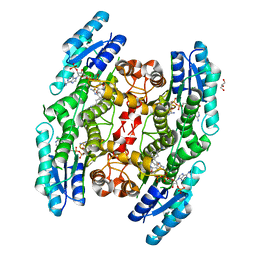 | | Trypanosoma brucei PTR1 in complex with inhibitor 1e (F206) | | Descriptor: | 4-[(2-azanyl-1,3-benzothiazol-6-yl)oxymethyl]benzenecarbonitrile, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2018-04-18 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
6GEX
 
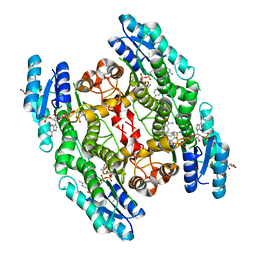 | | Trypanosoma brucei PTR1 in complex with inhibitor 2h (F246) | | Descriptor: | 4-[(2-azanyl-1,3-benzothiazol-6-yl)sulfanylmethyl]-~{N}-(phenylmethyl)benzamide, ACETATE ION, GLYCEROL, ... | | Authors: | Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
6GD4
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 4c (F188) | | Descriptor: | 2-amino-1,3-benzothiazole-6-carboxamide, ACETATE ION, GLYCEROL, ... | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2018-04-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
6GCQ
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 2b (F192) | | Descriptor: | 6-(trifluoromethylsulfanyl)-1,3-benzothiazol-2-amine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2018-04-18 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
6GCL
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 3a (F020) | | Descriptor: | 6-methylsulfonyl-1,3-benzothiazol-2-amine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2018-04-18 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
6GCP
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 2d (F186) | | Descriptor: | 6-[(3,4-dichlorophenyl)methylsulfanyl]-1,3-benzothiazol-2-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2018-04-18 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
6GDP
 
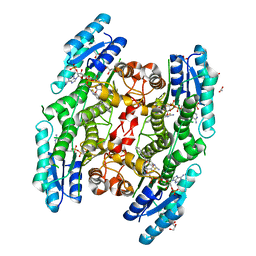 | | Trypanosoma brucei PTR1 in complex with inhibitor 4l (F162) | | Descriptor: | 2-azanyl-~{N}-(diphenylmethyl)-1,3-benzothiazole-6-carboxamide, ACETATE ION, GLYCEROL, ... | | Authors: | Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
7YRQ
 
 | | Cryo-EM structure of human Peroxisomal ABC Transporter ABCD1 | | Descriptor: | ATP-binding cassette sub-family D member 1, [(2~{R})-1-hexadecanoyloxy-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-propan-2-yl] octadec-9-enoate | | Authors: | Chao, X, Li-Na, J, Lin, T. | | Deposit date: | 2022-08-10 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into substrate recognition and translocation of human peroxisomal ABC transporter ALDP.
Signal Transduct Target Ther, 8, 2023
|
|
2BBD
 
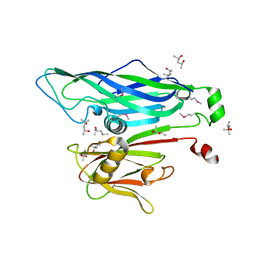 | | Crystal Structure of the STIV MCP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, coat protein | | Authors: | Khayat, R. | | Deposit date: | 2005-10-17 | | Release date: | 2005-12-06 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3T0H
 
 | |
3T0Z
 
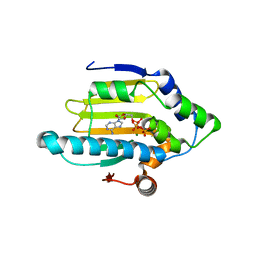 | | Hsp90 N-terminal domain bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
1M8S
 
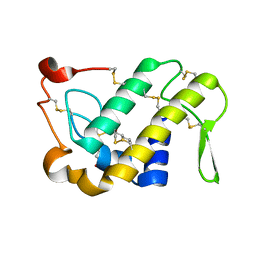 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 5.9) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase a2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
7XUP
 
 | | Crystal structure of TPe3.0 | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | Deposit date: | 2022-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
7XUQ
 
 | | Crystal structure of Tpe3.0 complexed with N-Boc-3-alkenylindole | | Descriptor: | Transcriptional regulator, PadR-like family, dimethyl 2-[[2-methyl-1-[(2-methylpropan-2-yl)oxycarbonyl]indol-3-yl]methyl]-2-prop-2-enyl-propanedioate | | Authors: | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | Deposit date: | 2022-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
2L6T
 
 | | Efficacy of an HIV-1 entry inhibitor targeting the GP41 fusion peptide | | Descriptor: | VIR-576 | | Authors: | Forssmann, W, The, Y, Stoll, M, Adermann, K, Albrecht, U, Barlos, K, Busmann, A, Can les-Mayordomo, A, Gimenez-Gallego, G, Hirsch, J, Jimenez-Barbero, J, Meyer-Olson, D, Muench, J, Perez-Castells, J, Staendker, L, Kirchhoff, F, Schmidt, R.E. | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Short-term monotherapy in HIV-infected patients with a virus entry inhibitor against the gp41 fusion peptide.
Sci Transl Med, 2, 2010
|
|
