3ONE
 
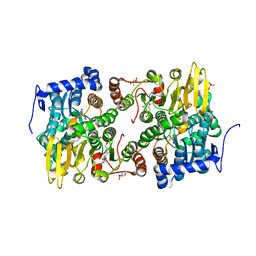 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3OND
 
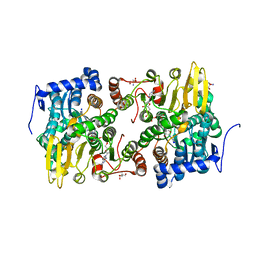 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4EWQ
 
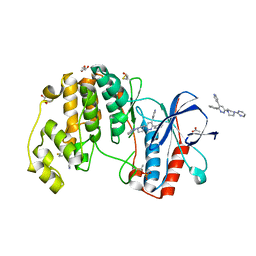 | | Human p38 alpha MAPK in complex with a pyridazine based inhibitor | | Descriptor: | 3-phenyl-4-(pyridin-4-yl)-6-[4-(pyrimidin-2-yl)piperazin-1-yl]pyridazine, 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, ACETATE ION, ... | | Authors: | Grum-Tokars, V, Minasov, G, Roy, S, Schavocky, J, Winsor, J, Watterson, D.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-27 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Selective and Brain Penetrant p38 alpha MAPK Inhibitor Candidate for Neurologic and Neuropsychiatric Disorders That Attenuates Neuroinflammation and Cognitive Dysfunction.
J.Med.Chem., 62, 2019
|
|
3ONF
 
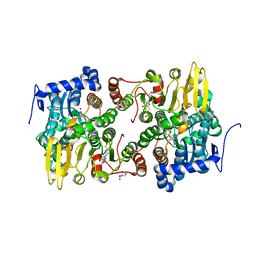 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with cordycepin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3'-DEOXYADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4RPV
 
 | | co-crystal structure of Pim1 with compound 3 | | Descriptor: | (3S)-1-{6-[5-(2,6-difluorophenyl)-2H-indazol-3-yl]pyrazin-2-yl}piperidin-3-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Huang, X. | | Deposit date: | 2014-10-31 | | Release date: | 2015-02-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The discovery of novel 3-(pyrazin-2-yl)-1H-indazoles as potent pan-Pim kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4MPY
 
 | | 1.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus (IDP00699) in complex with NAD+ | | Descriptor: | Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-14 | | Release date: | 2013-10-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus.
Appl.Environ.Microbiol., 80, 2014
|
|
4MPB
 
 | | 1.7 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus | | Descriptor: | Betaine aldehyde dehydrogenase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus.
Appl.Environ.Microbiol., 80, 2014
|
|
4JM7
 
 | | 1.82 Angstrom resolution crystal structure of holo-(acyl-carrier-protein) synthase (acpS) from Staphylococcus aureus | | Descriptor: | Holo-[acyl-carrier-protein] synthase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Structural characterization and comparison of three acyl-carrier-protein synthases from pathogenic bacteria.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4NEA
 
 | | 1.90 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, POTASSIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2M59
 
 | |
2MET
 
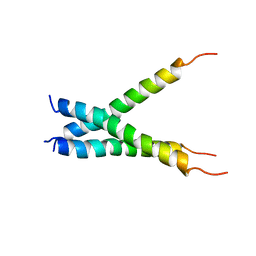 | | NMR spatial structure of the trimeric mutant TM domain of VEGFR2 receptor. | | Descriptor: | Vascular endothelial growth factor receptor 2 | | Authors: | Mineev, K.S, Arseniev, A.A, Shulepko, M, Lyukmanova, E.N, Kirpichnikov, M.P. | | Deposit date: | 2013-10-02 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of alternative transmembrane domain conformations in VEGF receptor 2 activation
Structure, 22, 2014
|
|
2MEU
 
 | | NMR spatial structure of mutant dimeric TM domain of VEGFR2 receptor | | Descriptor: | Vascular endothelial growth factor receptor 2 | | Authors: | Mineev, K.S, Arseniev, A.S, Lyukmanova, E.N, Shulepko, M.A, Kirpichnikov, M.P. | | Deposit date: | 2013-10-02 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of alternative transmembrane domain conformations in VEGF receptor 2 activation
Structure, 22, 2014
|
|
3QMN
 
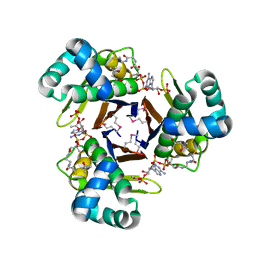 | | Crystal Structure of 4'-Phosphopantetheinyl Transferase AcpS from Vibrio cholerae O1 biovar eltor | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Kim, Y, Halavaty, A.S, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-04 | | Release date: | 2011-03-02 | | Last modified: | 2012-12-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization and comparison of three acyl-carrier-protein synthases from pathogenic bacteria.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3JS3
 
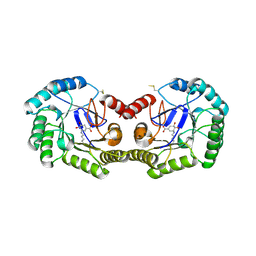 | | Crystal structure of type I 3-dehydroquinate dehydratase (aroD) from Clostridium difficile with covalent reaction intermediate | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, 3-dehydroquinate dehydratase | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-09 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.
J.Biol.Chem., 286, 2011
|
|
1JVD
 
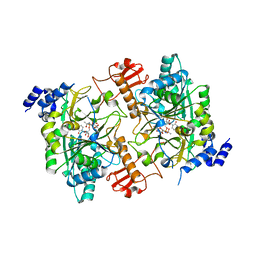 | | CRYSTAL STRUCTURE OF HUMAN AGX2 COMPLEXED WITH UDPGLCNAC | | Descriptor: | UDPGLCNAC PYROPHOSPHORYLASE (AGX2), URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Peneff, C, Bourne, Y. | | Deposit date: | 2001-08-29 | | Release date: | 2002-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture.
EMBO J., 20, 2001
|
|
1JVG
 
 | | CRYSTAL STRUCTURE OF HUMAN AGX2 COMPLEXED WITH UDPGALNAC | | Descriptor: | GLCNAC1P URIDYLTRANSFERASE, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | Peneff, C, Bourne, Y. | | Deposit date: | 2001-08-30 | | Release date: | 2002-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture.
EMBO J., 20, 2001
|
|
1JV3
 
 | | CRYSTAL STRUCTURE OF HUMAN AGX1 COMPLEXED WITH UDPGALNAC | | Descriptor: | GlcNAc1P uridyltransferase isoform 1: AGX1, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | Peneff, C, Bourne, Y. | | Deposit date: | 2001-08-28 | | Release date: | 2002-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture.
EMBO J., 20, 2001
|
|
1JV1
 
 | | CRYSTAL STRUCTURE OF HUMAN AGX1 COMPLEXED WITH UDPGLCNAC | | Descriptor: | GlcNAc1P uridyltransferase isoform 1: AGX1, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Peneff, C, Bourne, Y. | | Deposit date: | 2001-08-28 | | Release date: | 2002-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture.
EMBO J., 20, 2001
|
|
3ROH
 
 | | Crystal Structure of Leukotoxin (LukE) from Staphylococcus aureus subsp. aureus COL. | | Descriptor: | CHLORIDE ION, Leucotoxin LukEv, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the components of the Staphylococcus aureus leukotoxin ED.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
4LBT
 
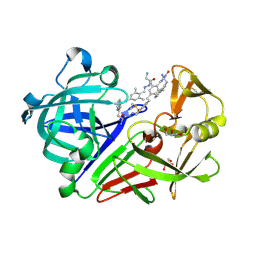 | | Endothiapepsin in complex with 100mM acylhydrazone inhibitor | | Descriptor: | (2S)-2-azanyl-3-(3H-indol-3-yl)-N-[(E)-(2,4,6-trimethylphenyl)methylideneamino]propanamide, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Radeva, N, Heine, A, Winquist, J, Klebe, G. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structure-based design of inhibitors of the aspartic protease endothiapepsin by exploiting dynamic combinatorial chemistry.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4MBO
 
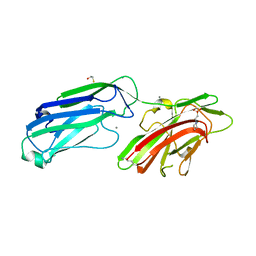 | | 1.65 Angstrom Crystal Structure of Serine-rich Repeat Adhesion Glycoprotein (Srr1) from Streptococcus agalactiae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
4MBR
 
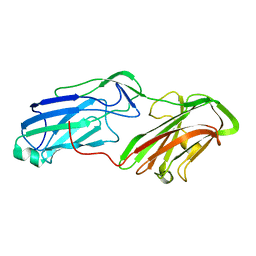 | | 3.65 Angstrom Crystal Structure of Serine-rich Repeat Protein (Srr2) from Streptococcus agalactiae | | Descriptor: | Serine-rich repeat protein 2 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
4MVJ
 
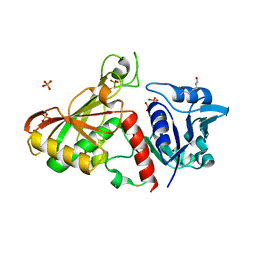 | | 2.85 Angstrom Resolution Crystal Structure of Glyceraldehyde 3-phosphate Dehydrogenase A (gapA) from Escherichia coli Modified by Acetyl Phosphate. | | Descriptor: | ACETATE ION, ACETYLPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Kuhn, M, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
Plos One, 9, 2014
|
|
4KMQ
 
 | | 1.9 Angstrom resolution crystal structure of uncharacterized protein lmo2446 from Listeria monocytogenes EGD-e | | Descriptor: | Lmo2446 protein, MAGNESIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E.V, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure to function of an alpha-glucan metabolic pathway that promotes Listeria monocytogenes pathogenesis.
Nat Microbiol, 2, 2016
|
|
4K6A
 
 | | Revised Crystal Structure of apo-form of Triosephosphate Isomerase (tpiA) from Escherichia coli at 1.8 Angstrom Resolution. | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Minasov, G, Kuhn, M, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
PLoS ONE, 9, 2014
|
|
