7EBV
 
 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in luteolin- and glutathione-bound form | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7E4D
 
 | | Crystal structure of PlDBR | | Descriptor: | Double Bond Reductase | | Authors: | Sugimoto, K, Senda, M, Senda, T. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploration and structure-based engineering of alkenal double bond reductases catalyzing the C alpha C beta double bond reduction of coniferaldehyde.
N Biotechnol, 68, 2022
|
|
7EBU
 
 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in Daidzein- and glutathione-bound form | | Descriptor: | 7-hydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
1FSK
 
 | | COMPLEX FORMATION BETWEEN A FAB FRAGMENT OF A MONOCLONAL IGG ANTIBODY AND THE MAJOR ALLERGEN FROM BIRCH POLLEN BET V 1 | | Descriptor: | ANTIBODY HEAVY CHAIN FAB, IMMUNOGLOBULIN KAPPA LIGHT CHAIN, MAJOR POLLEN ALLERGEN BET V 1-A | | Authors: | Mirza, O, Henriksen, A, Ipsen, H, Larsen, J, Wissenbach, M, Spangfort, M, Gajhede, M. | | Deposit date: | 2000-09-11 | | Release date: | 2000-10-02 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1.
J.Immunol., 165, 2000
|
|
3Q2S
 
 | | Crystal Structure of CFIm68 RRM/CFIm25 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Yang, Q, Coseno, M, Gilmartin, G.M, Doublie, S. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Human Cleavage Factor CFI(m)25/CFI(m)68/RNA Complex Provides an Insight into Poly(A) Site Recognition and RNA Looping.
Structure, 19, 2011
|
|
3Q2T
 
 | | Crystal Structure of CFIm68 RRM/CFIm25/RNA complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6, RNA | | Authors: | Yang, Q, Coseno, M, Gilmartin, G.M, Doublie, S. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.061 Å) | | Cite: | Crystal Structure of a Human Cleavage Factor CFI(m)25/CFI(m)68/RNA Complex Provides an Insight into Poly(A) Site Recognition and RNA Looping.
Structure, 19, 2011
|
|
4BAB
 
 | | Redesign of a Phenylalanine Aminomutase into a beta-Phenylalanine Ammonia Lyase | | Descriptor: | PHENYLALANINE AMINOMUTASE | | Authors: | Bartsch, S, Wybenga, G.G, Jansen, M, Heberling, M.M, Wu, B, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2012-09-12 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Redesign of a Phenylalanine Aminomutase Into a Phenylalanine Ammonia Lyase
To be Published
|
|
4BLF
 
 | | Variable internal flexibility characterizes the helical capsid formed by Agrobacterium VirE2 protein on single-stranded DNA. | | Descriptor: | SINGLE-STRAND DNA-BINDING PROTEIN | | Authors: | Bharat, T.A.M, Zbaida, D, Eisenstein, M, Frankenstein, Z, Mehlman, T, Weiner, L, Sorzano, C.O.S, Barak, Y, Albeck, S, Briggs, J.A.G, Wolf, S.G, Elbaum, M. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Variable Internal Flexibility Characterizes the Helical Capsid Formed by Agrobacterium Vire2 Protein on Single-Stranded DNA.
Structure, 21, 2013
|
|
1JQV
 
 | | The K213E mutant of Lactococcus lactis Dihydroorotate dehydrogenase A | | Descriptor: | ACETIC ACID, Dihydroorotate dehydrogenase A, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-09 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JRB
 
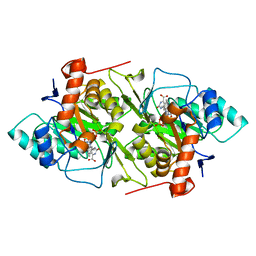 | | The P56A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, OROTIC ACID, dihydroorotate dehydrogenase A | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-13 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JRC
 
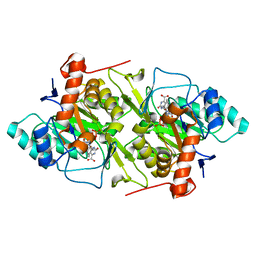 | | The N67A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, OROTIC ACID, dihydroorotate dehydrogenase A | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-13 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JUE
 
 | | 1.8 A resolution structure of native lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | ACETIC ACID, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-24 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
4BAA
 
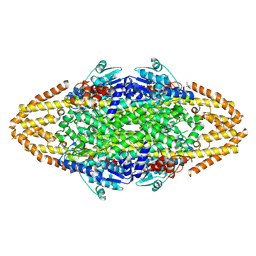 | | Redesign of a Phenylalanine Aminomutase into a beta-Phenylalanine Ammonia Lyase | | Descriptor: | PHENYLALANINE AMMONIA-LYASE | | Authors: | Bartsch, S, Wybenga, G.G, Jansen, M, Heberling, M.M, Wu, B, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2012-09-12 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Redesign of a Phenylalanine Aminomutase Into a Phenylalanine Ammonia Lyase
To be Published
|
|
1EK4
 
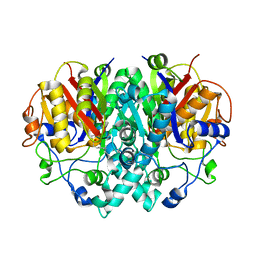 | | BETA-KETOACYL [ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH DODECANOIC ACID TO 1.85 RESOLUTION | | Descriptor: | BETA-KETOACYL [ACYL CARRIER PROTEIN] SYNTHASE I, LAURIC ACID | | Authors: | Olsen, J.G, Kadziola, A, Siggaard-Andersen, M, von Wettstein-Knowles, P, Larsen, S. | | Deposit date: | 2000-03-06 | | Release date: | 2001-04-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of beta-ketoacyl-acyl carrier protein synthase I complexed with fatty acids elucidate its catalytic machinery.
Structure, 9, 2001
|
|
4BEX
 
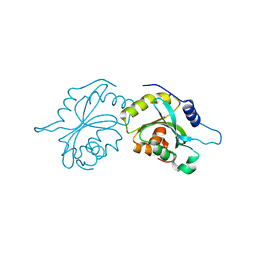 | | Structure of human Cofilin1 | | Descriptor: | COFILIN-1 | | Authors: | Klejnot, M, Gabrielsen, M, Cameron, J, Mleczak, A, Talapatra, S.K, Kozielski, F, Pannifer, A, Olson, M.F. | | Deposit date: | 2013-03-12 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the Human Cofilin1 Structure Reveals Conformational Changes Required for Actin-Binding
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1JUB
 
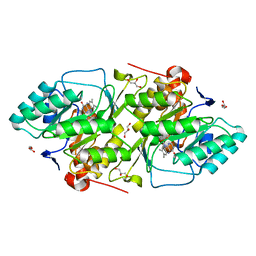 | | The K136E mutant of lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-24 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JQX
 
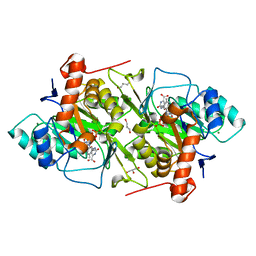 | | The R57A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-09 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1FN8
 
 | | FUSARIUM OXYSPORUM TRYPSIN AT ATOMIC RESOLUTION | | Descriptor: | GLY-ALA-ARG, GLYCEROL, SULFATE ION, ... | | Authors: | Rypniewski, W.R, Oestergaard, P, Noerregaard-Madsen, M, Dauter, M, Wilson, K.S. | | Deposit date: | 2000-08-21 | | Release date: | 2001-02-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: a study of ligand binding.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4EMS
 
 | |
3BFT
 
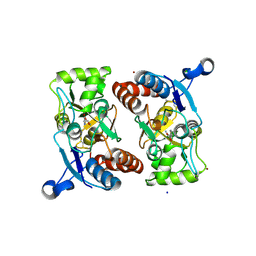 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (S)-TDPA at 2.25 A resolution | | Descriptor: | (2S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
1S88
 
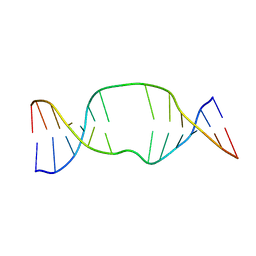 | | NMR structure of a DNA duplex with two INA nucleotides inserted opposite each other, dCTCAACXCAAGCT:dAGCTTGXGTTGAG | | Descriptor: | 5'-D(*AP*GP*CP*TP*TP*GP*(2DM)P*GP*TP*TP*GP*AP*G)-3', 5'-D(*CP*TP*CP*AP*AP*CP*(2DM)P*CP*AP*AP*GP*CP*T)-3' | | Authors: | Nielsen, C.B, Petersen, M, Pedersen, E.B, Hansen, P.E, Christensen, U.B. | | Deposit date: | 2004-01-31 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a modified DNA oligonucleotide containing a new intercalating nucleic acid.
Bioconjug.Chem., 15, 2004
|
|
1H0Q
 
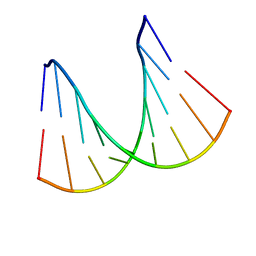 | | NMR solution structure of a fully modified locked nucleic acid (LNA) hybridized to RNA | | Descriptor: | 5-D(*(LKC)P*(TLN)P*(LCG)P*(LCA)P*(TLN)P*(LCA)P* (TLN)P*(LCG)P*(LCC))-3, 5-R(*GP*CP*AP*UP*AP*UP*CP*AP*G)-3 | | Authors: | Rasmussen, J, Petersen, M, Nielsen, K.E, Kumar, R, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2002-06-27 | | Release date: | 2003-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of Fully Modified Locked Nucleic Acid (Lna) Hybrids: Solution Structure of an Lna:RNA Hybrid and Characterization of an Lna:RNA Hybrid
Bioconjug.Chem., 15, 2004
|
|
2JIC
 
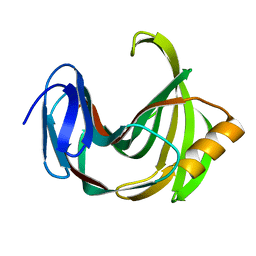 | | High resolution structure of xylanase-II from one micron beam experiment | | Descriptor: | XYLANASE-II | | Authors: | Moukhametzianov, R, Burghammer, M, Edwards, P.C, Petitdemange, S, Popov, D, Fransen, M, Schertler, G.F, Riekel, C. | | Deposit date: | 2007-02-27 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein Crystallography with a Micrometre-Sized Synchrotron-Radiation Beam.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2JRE
 
 | | C60-1, a PDZ domain designed using statistical coupling analysis | | Descriptor: | C60-1 PDZ domain peptide | | Authors: | Larson, C, Stiffler, M, Li, P, Rosen, M, MacBeath, G, Ranganathan, R. | | Deposit date: | 2007-06-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | C60-1, a PDZ domain designed using statistical coupling analysis
To be Published
|
|
1GV6
 
 | | Solution structure of alfa-L-LNA:DNA duplex | | Descriptor: | 5- D(*CP*(ATL)P*GP*CP*(ATL)P*(ATL)P*CP*(ATL)P* GP*C) -3, 5- D(*GP*CP*AP*GP*AP*AP*GP*CP*AP*G) -3 | | Authors: | Nielsen, K.M.E, Petersen, M, Haakansson, A.E, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alfa-L-Lna (Alfa-L-Ribo Configured Locked Nucleic Acid) Recognition of DNA: An NMR Spectroscopic Study
Chemistry, 8, 2002
|
|
