1X7Z
 
 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1X7Y
 
 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1X80
 
 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1X7W
 
 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
1X7X
 
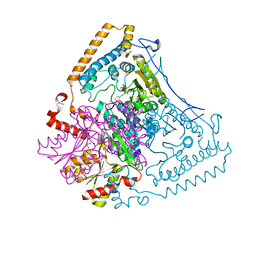 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, CHLORIDE ION, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-08-16 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
8IG5
 
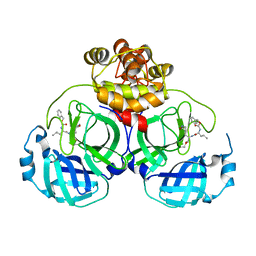 | | Crystal structure of SARS main protease in complex with GC376 | | Descriptor: | 3C-like proteinase nsp5, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IM6
 
 | | Crystal structure of HCoV 229E main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, Y.R, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of main proteases of HCoV-229E bound to inhibitor PF-07304814 and PF-07321332.
Biochem.Biophys.Res.Commun., 657, 2023
|
|
8IG4
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with GC376 | | Descriptor: | Non-structural protein 11, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG9
 
 | |
8IG8
 
 | |
8IG6
 
 | | Crystal structure of MERS main protease in complex with GC376 | | Descriptor: | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, ORF1a | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG7
 
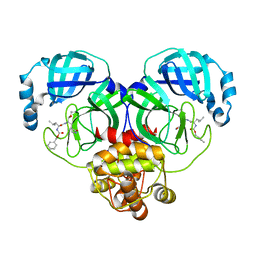 | |
3J9B
 
 | | Electron cryo-microscopy of an RNA polymerase | | Descriptor: | Polymerase, Polymerase basic protein 2, RNA (5'-R(*UP*UP*UP*UP*UP*A)-3'), ... | | Authors: | Chang, S.H, Sun, D.P, Liang, H.H, Wang, J, Li, J, Guo, L, Wang, X.L, Guan, C.C, Boruah, B.M, Yuan, L.M, Feng, F, Yang, M.R, Wojdyla, J, Wang, J.W, Wang, M.T, Wang, H.W, Liu, Y.F. | | Deposit date: | 2014-12-16 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM Structure of Influenza Virus RNA Polymerase Complex at 4.3 angstrom Resolution.
Mol.Cell, 2015
|
|
8IPT
 
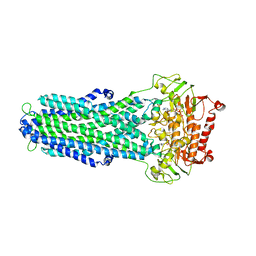 | |
8IPR
 
 | |
8IPS
 
 | |
8IPQ
 
 | |
1FXW
 
 | |
2QLD
 
 | | human Hsp40 Hdj1 | | Descriptor: | DnaJ homolog subfamily B member 1 | | Authors: | Hu, J, Wu, Y, Li, J, Fu, Z, Sha, B. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the putative peptide-binding fragment from the human Hsp40 protein Hdj1.
Bmc Struct.Biol., 8, 2008
|
|
5DP4
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 3 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2S)-2-methyl-3-phenylpropanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP9
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 9 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(cyclobutylmethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP8
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 8 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(2-cyclopropylethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DPA
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 6 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-acetyl-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP5
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 4 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2R,5S)-5-amino-2-(4-fluorobenzyl)-6-methyl-4-oxoheptanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2016-04-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP7
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 5 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-methylbutanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
