3E55
 
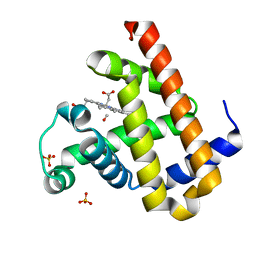 | | Carbonmonoxy Sperm Whale Myoglobin at 100 K: Laser off | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-08-13 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1DJU
 
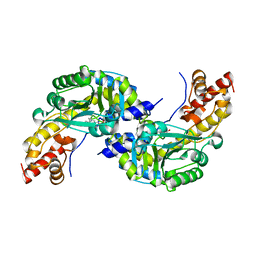 | | CRYSTAL STRUCTURE OF AROMATIC AMINOTRANSFERASE FROM PYROCOCCUS HORIKOSHII OT3 | | Descriptor: | AROMATIC AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matsui, I, Matsui, E, Sakai, Y, Kikuchi, H, Kawarabayashi, H. | | Deposit date: | 1999-12-06 | | Release date: | 2001-04-11 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular structure of hyperthermostable aromatic aminotransferase with novel substrate specificity from Pyrococcus horikoshii.
J.Biol.Chem., 275, 2000
|
|
1ITU
 
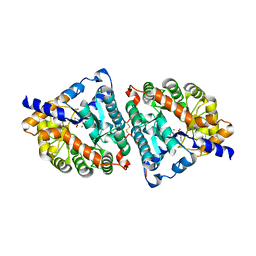 | | HUMAN RENAL DIPEPTIDASE COMPLEXED WITH CILASTATIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CILASTATIN, RENAL DIPEPTIDASE, ... | | Authors: | Nitanai, Y, Satow, Y, Adachi, H, Tsujimoto, M. | | Deposit date: | 2002-02-03 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Renal Dipeptidase Involved in beta-Lactam Hydrolysis
J.Mol.Biol., 321, 2002
|
|
1WW9
 
 | | Crystal structure of the terminal oxygenase component of carbazole 1,9a-dioxygenase, a non-heme iron oxygenase system catalyzing the novel angular dioxygenation for carbazole and dioxin | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, terminal oxygenase component of carbazole | | Authors: | Nojiri, H, Ashikawa, Y, Noguchi, H, Nam, J.-W, Urata, M, Fujimoto, Z, Mizuno, H, Yoshida, T, Habe, H, Omori, T. | | Deposit date: | 2005-01-05 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the terminal oxygenase component of angular dioxygenase, carbazole 1,9a-dioxygenase
J.Mol.Biol., 351, 2005
|
|
4YT1
 
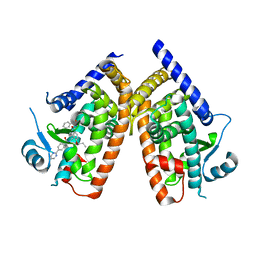 | | Human PPAR Gamma Ligand Binding Domain in complex with a Gammma Selective Synthetic Partial Agonist MEKT76 | | Descriptor: | N-(benzylsulfonyl)-4-propoxy-3-({[4-(pyrimidin-2-yl)benzoyl]amino}methyl)benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Ohashi, M, Miyachi, H, Kusunoki, M. | | Deposit date: | 2015-03-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peroxisome proliferator-activated receptor gamma (PPAR gamma ) has multiple binding points that accommodate ligands in various conformations: Structurally similar PPAR gamma partial agonists bind to PPAR gamma LBD in different conformations
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1V6Y
 
 | | Crystal Structure Of chimeric Xylanase between Streptomyces Olivaceoviridis E-86 FXYN and Cellulomonas fimi Cex | | Descriptor: | Beta-xylanase,Exoglucanase/xylanase | | Authors: | Kaneko, S, Ichinose, H, Fujimoto, Z, Kuno, A, Yura, K, Go, M, Mizuno, H, Kusakabe, I, Kobayashi, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-09-07 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of a family 10 beta-xylanase chimera of Streptomyces olivaceoviridis E-86 FXYN and Cellulomonas fimi Cex
J.Biol.Chem., 279, 2004
|
|
4ZCN
 
 | | Crystal structure of nvPizza2-S16S58 | | Descriptor: | IODIDE ION, SULFATE ION, nvPizza2-S16S58 | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of nvPizza2-S16S58
To Be Published
|
|
2D07
 
 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | Descriptor: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-07-26 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
4CUE
 
 | | Human Notch1 EGF domains 11-13 mutant T466V | | Descriptor: | CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7V5U
 
 | | The 0.92 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with 2-cyclohexadecylacetic acid (CYC16AA) | | Descriptor: | 2-cyclohexadecylethanoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-08-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | The 0.92 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with 2-cyclohexadecylacetic acid (CYC16AA)
To Be Published
|
|
7VB1
 
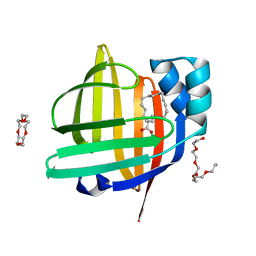 | | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with trans-vaccenic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-08-30 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with trans-vaccenic acid
To Be Published
|
|
4D0E
 
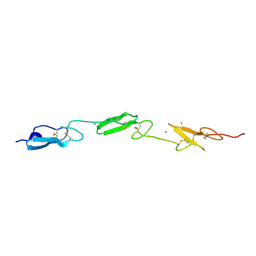 | | Human Notch1 EGF domains 11-13 mutant GlcNAc-fucose disaccharide modified at T466 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose, CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5C18
 
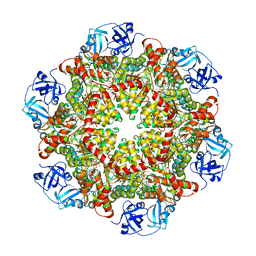 | | p97-delta709-728 in complex with ATP-gamma-S | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97.
Structure, 24, 2016
|
|
5C19
 
 | | p97 variant 2 in the apo state | | Descriptor: | SULFATE ION, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97.
Structure, 24, 2016
|
|
5C1B
 
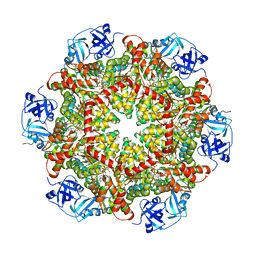 | |
2MX2
 
 | | UBX-L domain of VCIP135 | | Descriptor: | Deubiquitinating protein VCIP135 | | Authors: | Iwazu, T, Murayama, S, Igarashi, R, Hrioaki, H, Shirakawa, M, Tochio, H. | | Deposit date: | 2014-12-07 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and interaction mode of the UBX-L domain of VCIP135 determined by solution NMR spectroscopy
To be Published
|
|
5C1A
 
 | | p97-N750D/R753D/M757D/Q760D in complex with ATP-gamma-S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97.
Structure, 24, 2016
|
|
1UEL
 
 | | Solution structure of ubiquitin-like domain of hHR23B complexed with ubiquitin-interacting motif of proteasome subunit S5a | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, UV excision repair protein RAD23 homolog B | | Authors: | Fujiwara, K, Tenno, T, Jee, J.G, Sugasawa, K, Ohki, I, Kojima, C, Tochio, H, Hiroaki, H, Hanaoka, H, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-19 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ubiquitin-interacting Motif of S5a Bound to the Ubiquitin-like Domain of HR23B
J.Biol.Chem., 279, 2004
|
|
4CUD
 
 | | Human Notch1 EGF domains 11-13 mutant fucosylated at T466 | | Descriptor: | CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1, alpha-L-fucopyranose | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5CHB
 
 | | Crystal structure of nvPizza2-S16H58 coordinating a CdCl2 nanocrystal | | Descriptor: | CADMIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2015-07-10 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomineralization of a Cadmium Chloride Nanocrystal by a Designed Symmetrical Protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4CUF
 
 | | Human Notch1 EGF domains 11-13 mutant T466S | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7C7V
 
 | | Vitamin D3 receptor/lithochoric acid derivative complex | | Descriptor: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
7WJ1
 
 | | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with arachidonic acid | | Descriptor: | ARACHIDONIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with arachidonic acid
To Be Published
|
|
7WKB
 
 | | The 0.81 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with docosahexaenoic acid | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-01-08 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | The 0.81 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with docosahexaenoic acid
To Be Published
|
|
7WOM
 
 | | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with eicosapentaenoic acid | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Fatty acid-binding protein, heart | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-01-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with eicosapentaenoic acid
To Be Published
|
|
