6EPP
 
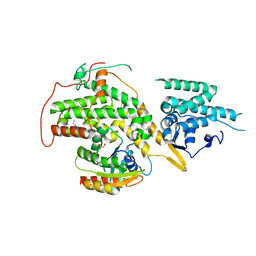 | | RAS GUANINE EXCHANGE FACTOR SOS1 (REM-CDC25) IN COMPLEX WITH KRAS(G12C) AND FRAGMENT SCREENING HIT F4 | | Descriptor: | GLYCEROL, GTPase KRas, Son of sevenless homolog 1, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Bader, B, Schroeder, J, Wortmann, L, Sautier, B, Kahmann, J, Wegener, D, Briem, H, Petersen, K, Badock, V. | | Deposit date: | 2017-10-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8UFH
 
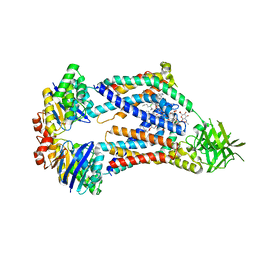 | | Acinetobacter baylyi LptB2FG bound to Acinetobacter baylyi lipopolysaccharide and a macrocyclic peptide | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-2-[(2~{R},4~{R},5~{R},6~{R})-5-[(2~{S},4~{R},5~{R},6~{R})-4-[(2~{R},3~{R},4~{R},5~{S},6~{S})-3-acetamido-6-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-5-oxidanyl-oxan-2-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{S})-3-dodecanoyloxydodecanoyl]oxy-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-heptanoyloxynonanoyl]amino]-3-oxidanyl-4-[(3~{R})-3-oxidanyloctanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-5-[[(3~{S})-3-[(3~{R})-3-oxidanyldodecanoyl]oxydecanoyl]amino]-3-phosphonooxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, (7S,10S,13S,17P)-10-(4-aminobutyl)-7-(3-aminopropyl)-17-(6-aminopyridin-3-yl)-20-chloro-13-[(1H-indol-3-yl)methyl]-12-methyl-6,7,9,10,12,13,15,16-octahydropyrido[2,3-b][1,5,8,11,14]benzothiatetraazacycloheptadecine-8,11,14(5H)-trione, LPS export ABC transporter permease LptG, ... | | Authors: | Pahil, K.S, Gilman, M.S.A, Baidin, V, Kruse, A.C, Kahne, D. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A new antibiotic traps lipopolysaccharide in its intermembrane transporter.
Nature, 625, 2024
|
|
8UFG
 
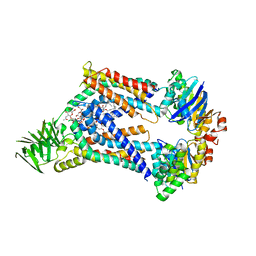 | | Acinetobacter baylyi LptB2FG bound to Acinetobacter baylyi lipopolysaccharide | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-2-[(2~{R},4~{R},5~{R},6~{R})-5-[(2~{S},4~{R},5~{R},6~{R})-4-[(2~{R},3~{R},4~{R},5~{S},6~{S})-3-acetamido-6-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-5-oxidanyl-oxan-2-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{S})-3-dodecanoyloxydodecanoyl]oxy-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-heptanoyloxyundecanoyl]amino]-3-oxidanyl-4-[(3~{R})-3-oxidanyloctanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-5-[[(3~{S})-3-[(3~{R})-3-oxidanyldecanoyl]oxydecanoyl]amino]-3-phosphonooxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, DODECYL-BETA-D-MALTOSIDE, LPS export ABC transporter permease LptG, ... | | Authors: | Pahil, K.S, Gilman, M.S.A, Baidin, V, Kruse, A.C, Kahne, D. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A new antibiotic traps lipopolysaccharide in its intermembrane transporter.
Nature, 625, 2024
|
|
9C29
 
 | | Hexadecamer of NL4-3 WT HIV-1 intasome | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*CP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*CP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*GP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*GP*CP*A)-3'), Integrase, ... | | Authors: | Lyumkis, D, Jing, T, Zhang, Z. | | Deposit date: | 2024-05-30 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Oligomeric HIV-1 Integrase Structures Reveal Functional Plasticity for Intasome Assembly and RNA Binding
Biorxiv, 2025
|
|
2R6F
 
 | | Crystal Structure of Bacillus stearothermophilus UvrA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Excinuclease ABC subunit A, ZINC ION | | Authors: | Inuzuka, Y, Pakotiprapha, D, Bowman, B.R, Jeruzalmi, D, Verdine, G.L. | | Deposit date: | 2007-09-05 | | Release date: | 2008-01-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Bacillus stearothermophilus UvrA Provides Insight into ATP-Modulated Dimerization, UvrB Interaction, and DNA Binding.
Mol.Cell, 29, 2008
|
|
6C2H
 
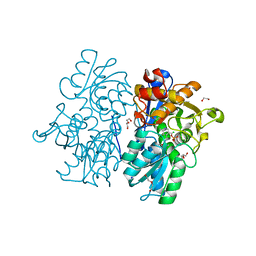 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the Catalytic Core | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
2X0R
 
 | | R207S, R292S Mutant of Malate Dehydrogenase from the Halophilic Archeon Haloarcula marismortui (HoloForm) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Irimia, A, Ebel, C, Vellieux, F.M.D, Richard, S.B, Cosenza, L.W, Zaccai, G, Madern, D. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.915 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
9D6J
 
 | | Nitrile hydratase BR52K mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Miller, C.G, Holz, R.C, Liu, D, Kaley, N. | | Deposit date: | 2024-08-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Role of second-sphere arginine residues in metal binding and metallocentre assembly in nitrile hydratases.
J Inorg Biochem, 256, 2024
|
|
9D6M
 
 | | Nitrile hydratase BR157K mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Miller, C.G, Holz, R.C, Liu, D, Kaley, N. | | Deposit date: | 2024-08-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Role of second-sphere arginine residues in metal binding and metallocentre assembly in nitrile hydratases.
J Inorg Biochem, 256, 2024
|
|
6C4P
 
 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PMP Complex | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Berkowitz, D.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
9D65
 
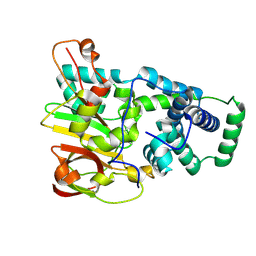 | | Nitrile hydratase BR52A mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Miller, C.G, Holz, R.C, Liu, D, Kaley, N. | | Deposit date: | 2024-08-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Role of second-sphere arginine residues in metal binding and metallocentre assembly in nitrile hydratases.
J Inorg Biochem, 256, 2024
|
|
9D6K
 
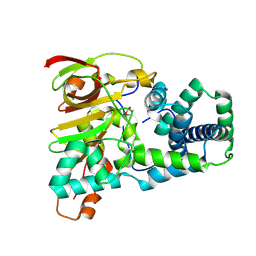 | | Nitrile hydratase BR157A mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, OXYGEN ATOM | | Authors: | Miller, C.G, Holz, R.C, Liu, D, Kaley, N. | | Deposit date: | 2024-08-15 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Role of second-sphere arginine residues in metal binding and metallocentre assembly in nitrile hydratases.
J Inorg Biochem, 256, 2024
|
|
7UEL
 
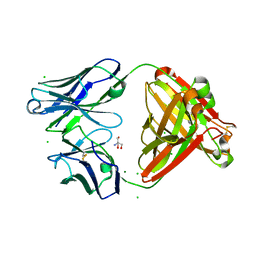 | |
7U0L
 
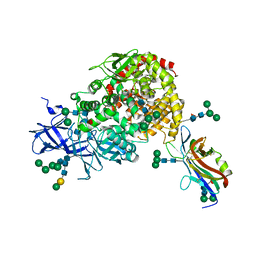 | | Crystal structure of the CCoV-HuPn-2018 RBD (domain B) in complex with canine APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7UEM
 
 | |
7UEN
 
 | |
7US9
 
 | | CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7USB
 
 | | CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7US6
 
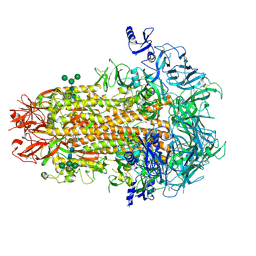 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7USF
 
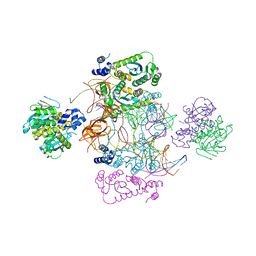 | |
7USA
 
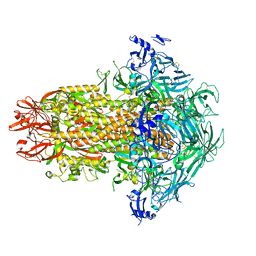 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
3NBY
 
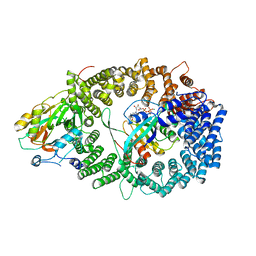 | | Crystal structure of the PKI NES-CRM1-RanGTP nuclear export complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8UAO
 
 | | DpHF18 filament | | Descriptor: | DpHF18 | | Authors: | Lynch, E.M, Shen, H, Kollman, J.M, Baker, D. | | Deposit date: | 2023-09-21 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 19, 2024
|
|
6RSR
 
 | | TBK1 in complex with compound 2 | | Descriptor: | Serine/threonine-protein kinase TBK1, ~{N}-(cyclopropen-1-ylmethyl)-2-[[4-[[4-[3,3,3-tris(fluoranyl)propanoyl]piperazin-1-yl]methyl]pyridin-2-yl]amino]-1~{H}-benzimidazole-5-carboxamide | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
7UCP
 
 | | computationally designed macrocycle | | Descriptor: | computationally designed cyclic peptide D8.3.p2 | | Authors: | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | Deposit date: | 2022-03-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
