3WN1
 
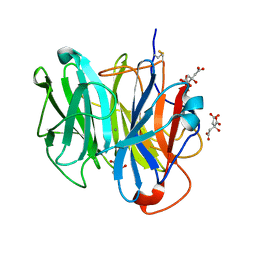 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with xylotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WN0
 
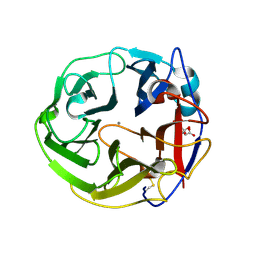 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with L-arabinose | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3VOR
 
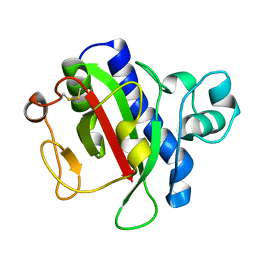 | | Crystal Structure Analysis of the CofA | | Descriptor: | CFA/III pilin | | Authors: | Fukakusa, S, Kawahara, K, Nakamura, S, Iwasita, T, Baba, S, Nishimura, M, Kobayashi, Y, Honda, T, Iida, T, Taniguchi, T, Ohkubo, T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-09-26 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VTE
 
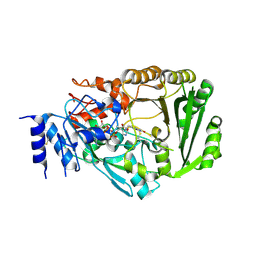 | | Crystal structure of tetrahydrocannabinolic acid synthase from Cannabis sativa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Tetrahydrocannabinolic acid synthase | | Authors: | Shoyama, Y, Tamada, T, Kurihara, K, Takeuchi, A, Taura, F, Arai, S, Blaber, M, Shoyama, Y, Morimoto, S, Kuroki, R. | | Deposit date: | 2012-05-28 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of 1-tetrahydrocannabinolic acid (THCA) synthase, the enzyme controlling the psychoactivity of Cannabis sativa
J.Mol.Biol., 423, 2012
|
|
2Z44
 
 | | Crystal Structure of Selenomethionine-labeled ORF134 | | Descriptor: | ORF134 | | Authors: | Tomimoto, Y, Ihara, K, Onizuka, T, Kanai, S, Ashida, H, Yokota, A, Tanaka, S, Miyasaka, H, Yamada, Y, Kato, R, Wakatsuki, S. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of ORF134
To be Published
|
|
2Z46
 
 | | Crystal Structure of Native-ORF134 | | Descriptor: | ORF134 | | Authors: | Tomimoto, Y, Ihara, K, Onizuka, T, Kanai, S, Ashida, H, Yokota, A, Tanaka, S, Miyasaka, H, Yamada, Y, Kato, R, Wakatsuki, S. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal Structure of ORF134
To be Published
|
|
2ZXY
 
 | | Crystal Structure of Cytochrome c555 from Aquifex aeolicus | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Obuchi, M, Kawahara, K, Motooka, D, Nakamura, S, Yamanaka, M, Takeda, T, Uchiyama, S, Kobayashi, Y, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Hyperstability and crystal structure of cytochrome c(555) from hyperthermophilic Aquifex aeolicus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2Z45
 
 | | Crystal Structure of Zn-bound ORF134 | | Descriptor: | ORF134, ZINC ION | | Authors: | Tomimoto, Y, Ihara, K, Onizuka, T, Kanai, S, Ashida, H, Yokota, A, Tanaka, S, Miyasaka, H, Yamada, Y, Kato, R, Wakatsuki, S. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of ORF134
To be Published
|
|
3B2C
 
 | | Crystal structure of the collagen triple helix model [{PRO-HYP(R)-GLY}4-{HYP(S)-Pro-GLY}2-{PRO-HYP(R)-GLY}4]3 | | Descriptor: | Collagen-like peptide | | Authors: | Motooka, D, Kawahara, K, Nakamura, S, Doi, M, Nishi, Y, Nishiuchi, Y, Nakazawa, T, Yoshida, T, Ohkubo, T, Kobayashi, Y, Kang, Y.K, Uchiyama, S. | | Deposit date: | 2011-07-26 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The triple helical structure and stability of collagen model peptide with 4(S)-hydroxyprolyl-pro-gly units
Biopolymers, 98, 2011
|
|
5YD8
 
 | |
8YFY
 
 | | CRYSTAL STRUCTURE OF THE EST1 H274D MUTANT AT PH 4.2 | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 2024
|
|
8YFZ
 
 | | CRYSTAL STRUCTURE OF THE EST1 H274E MUTANT AT PH 4.2 | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 2024
|
|
7XBQ
 
 | |
2RQP
 
 | |
5XI9
 
 | |
5XIR
 
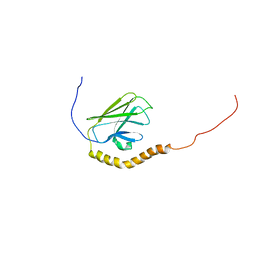 | |
4P9N
 
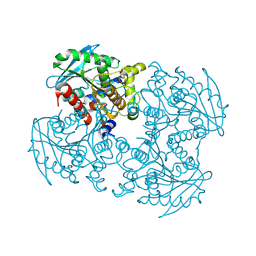 | | Crystal structure of sshesti PE mutant | | Descriptor: | Carboxylesterase | | Authors: | Unno, H. | | Deposit date: | 2014-04-04 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Low pH Adaptation of a Unique Carboxylesterase from Ferroplasma: ALTERING THE pH OPTIMA OF TWO CARBOXYLESTERASES.
J.Biol.Chem., 289, 2014
|
|
2DXS
 
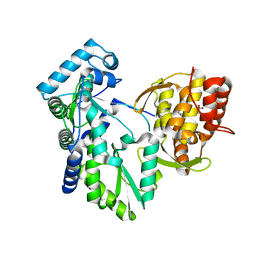 | | Crystal structure of HCV NS5B RNA polymerase complexed with a tetracyclic inhibitor | | Descriptor: | Genome polyprotein, N-[(13-CYCLOHEXYL-6,7-DIHYDROINDOLO[1,2-D][1,4]BENZOXAZEPIN-10-YL)CARBONYL]-2-METHYL-L-ALANINE | | Authors: | Adachi, T, Tsuruha, J, Doi, S, Murase, K, Ikegashira, K, Watanabe, S, Uehara, K, Orita, T, Nomura, A, Kamada, M. | | Deposit date: | 2006-08-30 | | Release date: | 2006-12-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Conformationally Constrained Tetracyclic Compounds as Potent Hepatitis C Virus NS5B RNA Polymerase Inhibitors
J.Med.Chem., 49, 2006
|
|
6M4B
 
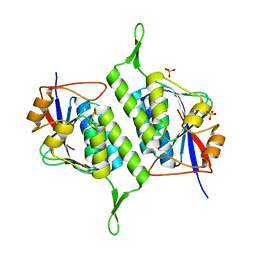 | |
7V5N
 
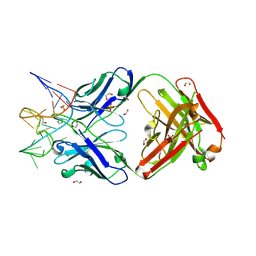 | | Crystal structure of Fab fragment of bevacizumab bound to DNA aptamer | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*GP*TP*TP*GP*GP*TP*GP*GP*TP*AP*GP*TP*TP*AP*CP*GP*TP*TP*CP*GP*C)-3'), IMIDAZOLE, ... | | Authors: | Hishiki, A, Tong, J, Todoroki, K, Hashimoto, H. | | Deposit date: | 2021-08-17 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of a DNA aptamer that binds to the complementarity-determining region of therapeutic monoclonal antibody and affinity improvement induced by pH-change for sensitive detection.
Biosens.Bioelectron., 203, 2022
|
|
5ZO9
 
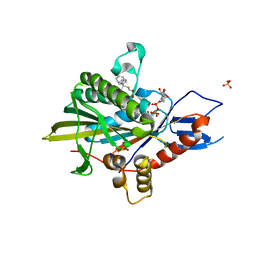 | | Eg5 motor domain in complex with STLC-type inhibitor PVEI0021 (C2 type) | | Descriptor: | (2R)-2-azanyl-3-[(4-methoxyphenyl)-diphenyl-methyl]sulfanyl-propanoic acid, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yokoyama, H, Sato, K. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Thermodynamic Basis of the Enhanced Interaction between Kinesin Spindle Protein Eg5 and STLC-type Inhibitors.
Acs Omega, 3, 2018
|
|
5ZO8
 
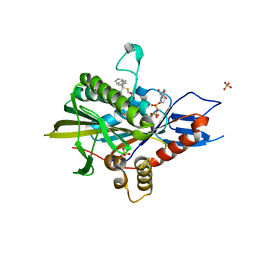 | | Eg5 motor domain in complex with STLC-type inhibitor PVEI0021 (P21 type) | | Descriptor: | (2R)-2-azanyl-3-[(4-methoxyphenyl)-diphenyl-methyl]sulfanyl-propanoic acid, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yokoyama, H, Sato, K. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Thermodynamic Basis of the Enhanced Interaction between Kinesin Spindle Protein Eg5 and STLC-type Inhibitors.
Acs Omega, 3, 2018
|
|
5ZO7
 
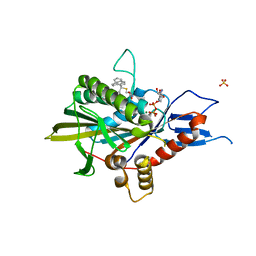 | | Kinesin spindle protein Eg5 in complex with STLC-type inhibitor PVEI0138 | | Descriptor: | (2R)-2-azanyl-3-[[2-(4-methoxyphenyl)-2-tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3,5,7,12,14-hexaenyl]sulfanyl]propanoic acid, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yokoyama, H, Sato, K. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Thermodynamic Basis of the Enhanced Interaction between Kinesin Spindle Protein Eg5 and STLC-type Inhibitors.
Acs Omega, 3, 2018
|
|
8HW1
 
 | | Far-red light-harvesting complex of Antarctic alga Prasiola crispa | | Descriptor: | (1S)-4-[(1E,3Z,5E,7E,9E,11E,13E,15E,17E)-3-(hydroxymethyl)-7,12,16-trimethyl-18-[(1R,4S)-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]-3,5,5-trimethyl-cyclohex-3-en-1-ol, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, CHLOROPHYLL A, ... | | Authors: | Kosugi, M, Kawasaki, M, Shibata, Y, Moriya, T, Adachi, N, Senda, T. | | Deposit date: | 2022-12-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Uphill energy transfer mechanism for photosynthesis in an Antarctic alga.
Nat Commun, 14, 2023
|
|
4XZF
 
 | | Crystal structure of HIRAN domain of human HLTF in complex with DNA | | Descriptor: | (DA)(DC)(DC)(DG)(DC)(DC)(DG)(DG)(DG)(DT)(DG)(DC)(DC), Helicase-like transcription factor | | Authors: | Hishiki, A, Hashimoto, H. | | Deposit date: | 2015-02-04 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of a Novel DNA-binding Domain of Helicase-like Transcription Factor (HLTF) and Its Functional Implication in DNA Damage Tolerance
J.Biol.Chem., 290, 2015
|
|
