2IAO
 
 | | Crystal structure of squid ganglion DFPase E37Q mutant | | Descriptor: | CALCIUM ION, DIISOPROPYLFLUOROPHOSPHATASE | | Authors: | Scharff, E.I, Koepke, J, Fritzsch, G, Luecke, C, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris
Structure, 9, 2001
|
|
2IAV
 
 | | Crystal structure of squid ganglion DFPase H287A mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Katsemi, V, Luecke, C, Koepke, J, Loehr, F, Maurer, S, Fritzsch, G, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Mutational and structural studies of the diisopropylfluorophosphatase from Loligo vulgaris shed new light on the catalytic mechanism of the enzyme
Biochemistry, 44, 2005
|
|
4UYF
 
 | | N-Terminal bromodomain of Human BRD2 with I-BET726 (GSK1324726A) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2S,4R)-1-acetyl-4-[(4-chlorophenyl)amino]-2-methyl-1,2,3,4-tetrahydroquinolin-6-yl]benzoic acid, BROMODOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Gosmini, R. | | Deposit date: | 2014-08-31 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Discovery of I-Bet726 (Gsk1324726A), a Potent Tetrahydroquinoline Apoa1 Up-Regulator and Selective Bet Bromodomain Inhibitor.
J.Med.Chem., 57, 2014
|
|
4UW7
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber without its intra-molecular chaperone domain | | Descriptor: | GLYCEROL, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
2IAT
 
 | | Crystal structure of squid ganglion DFPase W244L mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Scharff, E.I, Koepke, J, Fritzsch, G, Luecke, C, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris
Structure, 9, 2001
|
|
4WQB
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - bisulfite soak | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEME C, IODIDE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5013 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
8HT7
 
 | | The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*TP*GP*GP*G)-3'), GLN-ALA-GLN-ALA-THR-ILE-SER-PHE-PRO-LYS-ARG-LYS-LEU-SER-TRP | | Authors: | Liu, C, Zhu, G, Geng, Y, Xu, N. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex.
Int.J.Biol.Macromol., 260, 2024
|
|
4WQ7
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - "as isolated" form | | Descriptor: | HEME C, IODIDE ION, SULFATE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
2IWA
 
 | | Unbound glutaminyl cyclotransferase from Carica papaya. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLUTAMINE CYCLOTRANSFERASE, ... | | Authors: | Guevara, T, Mallorqui-Fernandez, N, Garcia-Castellanos, R, Petersen, G.E, Lauritzen, C, Pedersen, J, Arnau, J, Gomis-Ruth, F.X, Sola, M. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Papaya Glutamine Cyclotransferase Shows a Singular Five-Fold Beta-Propeller Architecture that Suggests a Novel Reaction Mechanism.
Biol.Chem., 387, 2006
|
|
6EWF
 
 | |
6F26
 
 | | Crystal structure of human Casein Kinase I delta in complex with compound 31b | | Descriptor: | (9~{S},10~{S},11~{R})-~{N}-[4-[3-(4-fluorophenyl)-5-propan-2-yl-1,2-oxazol-4-yl]pyridin-2-yl]-4-(4-methoxyphenyl)-10,11-bis(oxidanyl)-1,7-diazatricyclo[7.3.0.0^{3,7}]dodeca-3,5-diene-6-carboxamide, Casein kinase I isoform delta, SULFATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2017-11-23 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design, Synthesis and Biological Evaluation of Isoxazole-Based CK1 Inhibitors Modified with Chiral Pyrrolidine Scaffolds.
Molecules, 24, 2019
|
|
8I5N
 
 | | Rat Kir4.1 in complex with PIP2 and Lys05 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
2IXH
 
 | | RmlC P aeruginosa with dTDP-rhamnose | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RmlC, a C3' and C5' carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation.
J. Mol. Biol., 365, 2007
|
|
8I5M
 
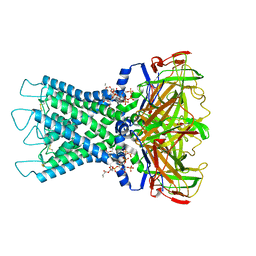 | | Rat Kir4.1 in complex with PIP2 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
6EU5
 
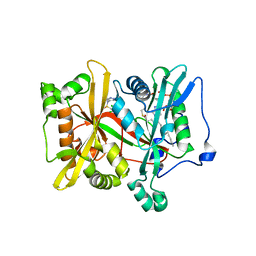 | | Leishmania major N-myristoyltransferase with bound myristoyl-CoA and inhibitor | | Descriptor: | 4-[3-[(8~{a}~{R})-3,4,6,7,8,8~{a}-hexahydro-1~{H}-pyrrolo[1,2-a]pyrazin-2-yl]propyl]-2,6-bis(chloranyl)-~{N}-methyl-~{N}-(1,3,5-trimethylpyrazol-4-yl)benzenesulfonamide, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA | | Authors: | Brenk, R, Kehrein, J, Kersten, C. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.496083 Å) | | Cite: | How To Design Selective Ligands for Highly Conserved Binding Sites: A Case Study UsingN-Myristoyltransferases as a Model System.
J.Med.Chem., 2019
|
|
8I4U
 
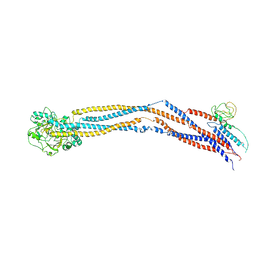 | | Cryo-EM structure of 5-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Qian, L, Jun, Z, Xiang, Z, Wang, Z, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2IFC
 
 | |
8I4X
 
 | | Cryo-EM structure of 5-subunit Smc5/6 | | Descriptor: | E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 5, Nse6, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HQS
 
 | | Cryo-EM structure of 8-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Wang, Z, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I21
 
 | | Cryo-EM structure of 6-subunit Smc5/6 arm region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Jun, Z, Qian, L, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2023-01-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.02 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2IXL
 
 | | RmlC S. suis with dTDP-rhamnose | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, NICKEL (II) ION | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
2IXC
 
 | | RmlC M. tuberculosis with dTDP-rhamnose | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE RMLC | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-07 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
1UFK
 
 | | Crystal structure of TT0836 | | Descriptor: | TT0836 protein | | Authors: | Kaminishi, T, Sakai, H, Takemoto-Hori, C, Terada, T, Nakagawa, N, Maoka, N, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-31 | | Release date: | 2003-11-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of TT0836
To be Published
|
|
6F1W
 
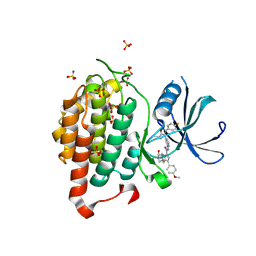 | | Crystal structure of human Casein Kinase I delta in complex with compound 31a | | Descriptor: | (9~{R},10~{R},11~{S})-~{N}-[4-[3-(4-fluorophenyl)-5-propan-2-yl-1,2-oxazol-4-yl]pyridin-2-yl]-4-(4-methoxyphenyl)-10,11-bis(oxidanyl)-1,7-diazatricyclo[7.3.0.0^{3,7}]dodeca-3,5-diene-6-carboxamide, Casein kinase I isoform delta, SULFATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2017-11-23 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Design, Synthesis and Biological Evaluation of Isoxazole-Based CK1 Inhibitors Modified with Chiral Pyrrolidine Scaffolds.
Molecules, 24, 2019
|
|
9LNP
 
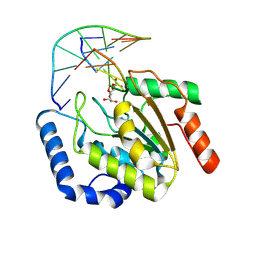 | | the hUNG bound to DNA product embedding uridine ribonucleotide | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*T*(RP5)P*AP*TP*CP*TP*T)-3'), ... | | Authors: | Liu, Y, Zhou, C, Zhan, X, Fan, C. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Uridine Embedded within DNA is Repaired by Uracil DNA Glycosylase via a Mechanism Distinct from That of Ribonuclease H2.
J.Am.Chem.Soc., 147, 2025
|
|
