7N7H
 
 | | X-ray crystal structure of Viperin-like enzyme from Nematostella vectensis | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, ... | | Authors: | Grove, T.L, Almo, S.C, Bonanno, J.B, Lachowicz, J.C, Gizzi, A.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Insight into the Substrate Scope of Viperin and Viperin-like Enzymes from Three Domains of Life.
Biochemistry, 60, 2021
|
|
5C5I
 
 | | Crystal structure of NADP-dependent dehydrogenase from Rhodobacter sphaeroides | | Descriptor: | NADP-dependent dehydrogenase | | Authors: | Kowiel, M, Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Porebski, P.J, Cymborowski, M, Al Obaidi, N.F, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of NADP-dependent dehydrogenase from Rhodobacter sphaeroides
to be published
|
|
5C7H
 
 | | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH | | Descriptor: | Aldo-keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-24 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH
to be published
|
|
5DM3
 
 | | Crystal Structure of Glutamine Synthetase from Chromohalobacter salexigens DSM 3043(Csal_0679, TARGET EFI-550015) with bound ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-glutamine synthetase | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Glutamine Synthetase from Chromohalobacter salexigens DSM 3043(Csal_0679, TARGET EFI-550015) with bound ADP
To be published
|
|
5E5M
 
 | | Crystal structure of mouse CTLA-4 in complex with nanobody | | Descriptor: | CTLA-4 nanobody, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-08 | | Release date: | 2016-10-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Crystal structure of mouse CTLA-4 in complex with nanobody
To Be Published
|
|
5CI5
 
 | | Crystal Structure of an ABC transporter Solute Binding Protein from Thermotoga Lettingae TMO (Tlet_1705, TARGET EFI-510544) bound with alpha-D-Tagatose | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein family 1, PENTAETHYLENE GLYCOL, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of an ABC transporter Solute Binding Protein from Thermotoga Lettingae TMO (Tlet_1705, TARGET EFI-510544) bound with alpha-D-Tagatose
To be published
|
|
5BXY
 
 | | Crystal structure of RNA methyltransferase from Salinibacter ruber in complex with S-Adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA methyltransferase, ... | | Authors: | Handing, K.B, LaRowe, C, Shabalin, I.G, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of RNA methylase family protein from Salinibacterruber in complex with S-Adenosyl-L-homocysteine.
to be published
|
|
7N2W
 
 | | The crystal structure of an FMN-dependent NADH-azoreductase, AzoA in complex with Red 40 | | Descriptor: | 6-hydroxy-5-[(E)-(2-methoxy-5-methyl-4-sulfophenyl)diazenyl]naphthalene-2-sulfonic acid, FLAVIN MONONUCLEOTIDE, FMN dependent NADH:quinone oxidoreductase | | Authors: | Arcinas, A.J, Fedorov, E, Kelly, L, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Uncovering a novel mechanism of enzyme activation in multimeric azoreductases
To Be Published
|
|
7N2X
 
 | | The crystal structure of an FMN-dependent NADH:quinone oxidoreductase, AzoR from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-AMINO-ACRYLIC ACID, ... | | Authors: | Arcinas, A.J, Fedorov, E, Kelly, L, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-05-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Uncovering a novel mechanism of enzyme activation in multimeric azoreductases
To Be Published
|
|
5E83
 
 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT440, CO-CRYSTALLIZATION EXPERIMENT | | Descriptor: | 2-methyl-3-({2-[1-(propan-2-yl)-1H-pyrazol-5-yl]pyridin-3-yl}methoxy)phenol, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Patskovska, L, Patskovsky, Y, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GBT440 increases haemoglobin oxygen affinity, reduces sickling and prolongs RBC half-life in a murine model of sickle cell disease.
Br.J.Haematol., 175, 2016
|
|
2DW6
 
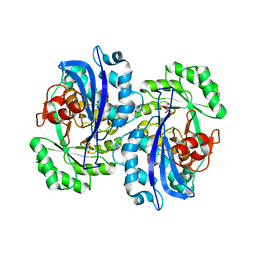 | | Crystal structure of the mutant K184A of D-Tartrate Dehydratase from Bradyrhizobium japonicum complexed with Mg++ and D-tartrate | | Descriptor: | Bll6730 protein, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium japonicum
Biochemistry, 45, 2006
|
|
2DW7
 
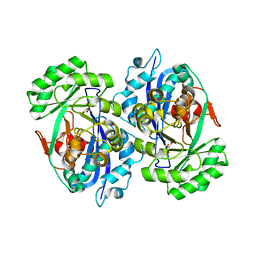 | | Crystal structure of D-tartrate dehydratase from Bradyrhizobium japonicum complexed with Mg++ and meso-tartrate | | Descriptor: | Bll6730 protein, MAGNESIUM ION, S,R MESO-TARTARIC ACID | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium japonicum
Biochemistry, 45, 2006
|
|
2F7F
 
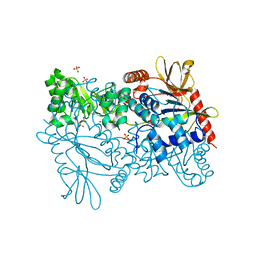 | | Crystal structure of Enterococcus faecalis putative nicotinate phosphoribosyltransferase, NEW YORK STRUCTURAL GENOMICS CONSORTIUM | | Descriptor: | DIPHOSPHATE, GLYCEROL, NICOTINIC ACID, ... | | Authors: | Patskovsky, Y, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Enterococcus Faecalis Nicotinate Phosphoribosyltransferase
To be Published
|
|
2FIQ
 
 | |
2FGS
 
 | | Crystal structure of Campylobacter jejuni YCEI protein, structural genomics | | Descriptor: | Putative periplasmic protein, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-22 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Campylobacter Jejuni YceI Periplasmic Protein
To be Published
|
|
2FLI
 
 | | The crystal structure of D-ribulose 5-phosphate 3-epimerase from Streptococus pyogenes complexed with D-xylitol 5-phosphate | | Descriptor: | D-XYLITOL-5-PHOSPHATE, ZINC ION, ribulose-phosphate 3-epimerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Akana, J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | d-Ribulose 5-Phosphate 3-Epimerase: Functional and Structural Relationships to Members of the Ribulose-Phosphate Binding (beta/alpha)(8)-Barrel Superfamily(,).
Biochemistry, 45, 2006
|
|
2G64
 
 | | Structure of Caenorhabditis elegans 6-pyruvoyl tetrahydropterin synthase | | Descriptor: | GLYCEROL, Putative 6-pyruvoyl tetrahydrobiopterin synthase, SODIUM ION, ... | | Authors: | Wengerter, B.C, Patskovsky, Y, Zhan, C, Ramagopal, U, Milstein, S, Vidal, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-02-24 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans 6-Pyruvoyl Tetrahydropterin Synthase
To be Published
|
|
2GDQ
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution | | Descriptor: | yitF | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-16 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution
To be Published
|
|
2GGE
 
 | | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, yitF | | Authors: | Malashkevich, V.N, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A
To be Published
|
|
2GL5
 
 | | Crystal Structure of Putative Dehydratase from Salmonella Thyphimurium | | Descriptor: | GLYCEROL, MAGNESIUM ION, putative dehydratase protein | | Authors: | Patskovsky, Y, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-04 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Putative Dehydratase from Salmonella Thyphimurium Lt2
To be Published
|
|
2GSH
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium | | Descriptor: | GLYCEROL, L-RHAMNONATE DEHYDRATASE, MAGNESIUM ION | | Authors: | Patskovsky, Y, Malashkevich, V.N, Sauder, J.M, Dickey, M, Adams, J.M, Wasserman, S.R, Gerlt, J, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal Structure of L-rhamnonate dehydratase from Salmonella Typhimurium Lt2
To be Published
|
|
2GQF
 
 | | Crystal structure of flavoprotein HI0933 from Haemophilus influenzae Rd | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Hypothetical protein HI0933, SULFATE ION | | Authors: | Mulichak, A.M, Patskovsky, Y, Keefe, L.J, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical flavoprotein HI0933 from Haemophilus influenzae Rd.
To be Published
|
|
2HC9
 
 | | Structure of Caenorhabditis elegans leucine aminopeptidase-zinc complex (LAP1) | | Descriptor: | BICARBONATE ION, GLYCEROL, Leucine aminopeptidase 1, ... | | Authors: | Zhan, C, Patskovsky, Y, Wengerter, B.C, Ramagopal, U, Milstein, S, Vidal, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-15 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure and Function of Caenorhabditis Elegans Leucine Aminopeptidase
To be Published
|
|
3LGX
 
 | |
1NW4
 
 | | Crystal Structure of Plasmodium falciparum Purine Nucleoside Phosphorylase in complex with ImmH and Sulfate | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Shi, W, Ting, L.M, Kicska, G.A, Lewandowicz, A, Tyler, P.C, Evans, G.B, Furneaux, R.H, Kim, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-02-05 | | Release date: | 2004-03-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plasmodium falciparum Purine Nucleoside Phosphorylase: CRYSTAL STRUCTURES, IMMUCILLIN INHIBITORS, AND DUAL CATALYTIC FUNCTION.
J.Biol.Chem., 279, 2004
|
|
