3EVN
 
 | | CRYSTAL STRUCTURE OF putative oxidoreductase from Streptococcus agalactiae 2603V/r | | Descriptor: | Oxidoreductase, Gfo/Idh/MocA family | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF putative Gfo/Idh/MocA family oxidoreductase from Streptococcus agalactiae
2603V/r
To be Published
|
|
3F2C
 
 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP and Mn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3F2D
 
 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP, Mn and Zn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3F7R
 
 | |
3FAY
 
 | | Crystal structure of the GAP-related domain of IQGAP1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ras GTPase-activating-like protein IQGAP1 | | Authors: | Kurella, V.B, Richard, J.M, Parke, C.L, Bellamy, H, Worthylake, D.K. | | Deposit date: | 2008-11-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the GTPase-activating protein-related domain from IQGAP1.
J.Biol.Chem., 284, 2009
|
|
3EYE
 
 | | Crystal structure of PTS system N-acetylgalactosamine-specific IIB component 1 from Escherichia coli | | Descriptor: | PTS system N-acetylgalactosamine-specific IIB component 1 | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Do, J, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of PTS system N-acetylgalactosamine-specific IIB component 1 from Escherichia coli
To be Published
|
|
3FK9
 
 | | Crystal structure of mMutator MutT protein from Bacillus halodurans | | Descriptor: | Mutator MutT protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of mMutator MutT protein from Bacillus halodurans
To be Published
|
|
3FBT
 
 | | Crystal structure of a chorismate mutase/shikimate 5-dehydrogenase fusion protein from Clostridium acetobutylicum | | Descriptor: | SULFATE ION, chorismate mutase and shikimate 5-dehydrogenase fusion protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a chorismate mutase/shikimate 5-dehydrogenase fusion protein from Clostridium acetobutylicum
To be Published
|
|
3FDW
 
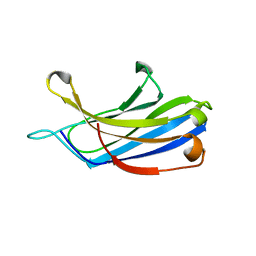 | | Crystal structure of a C2 domain from human synaptotagmin-like protein 4 | | Descriptor: | Synaptotagmin-like protein 4 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-26 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a C2 domain from human synaptotagmin-like protein 4
To be Published
|
|
3EYP
 
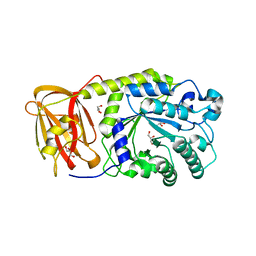 | | Crystal structure of putative alpha-L-fucosidase from Bacteroides thetaiotaomicron | | Descriptor: | GLYCEROL, Putative alpha-L-fucosidase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative alpha-L-fucosidase from Bacteroides thetaiotaomicron
To be Published
|
|
3F2B
 
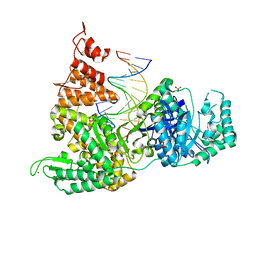 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP, Mg and Zn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3F7P
 
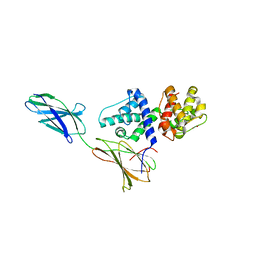 | |
3F7Q
 
 | |
3EVW
 
 | | Crystal structure of the Mimivirus NDK R107G mutant complexed with dTDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-10-13 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3EVY
 
 | | Crystal structure of a fragment of a putative type I restriction enzyme R protein from Bacteroides fragilis | | Descriptor: | Putative type I restriction enzyme R protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a fragment of a putative type I restriction enzyme R protein from Bacteroides fragilis
To be Published
|
|
3F13
 
 | | Crystal structure of putative nudix hydrolase family member from Chromobacterium violaceum | | Descriptor: | putative nudix hydrolase family member | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative nudix hydrolase family member from Chromobacterium violaceum
To be Published
|
|
3FCW
 
 | | Crystal structure of the Mimivirus NDK N62L mutant complexed with UDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-11-23 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3FBB
 
 | | Crystal structure of the Mimivirus NDK N62L-R107G double mutant complexed with UDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-11-19 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3FMY
 
 | |
3F1C
 
 | | CRYSTAL STRUCTURE OF 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Listeria monocytogenes | | Descriptor: | Putative 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase 2 | | Authors: | Patskovsky, Y, Ho, J, Toro, R, Gilmore, M, Miller, S, Groshong, C, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Listeria monocytogenes
To be Published
|
|
3F5S
 
 | | CRYSTAL STRUCTURE OF putatitve short chain dehydrogenase from Shigella flexneri 2a str. 301 | | Descriptor: | dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CRYSTAL STRUCTURE OF putatitve short chain dehydrogenase from Shigella flexneri 2a str. 301
To be Published
|
|
3F6C
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, Positive transcription regulator evgA | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Wu, B, Bain, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI
To be Published
|
|
3FHL
 
 | | Crystal structure of a putative oxidoreductase from bacteroides fragilis nctc 9343 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative oxidoreductase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-09 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of a Putative Oxidoreductase from Bacteroides Fragilis Nctc 9343
To be Published
|
|
7B8W
 
 | | Structure of LIMK1 Kinase domain with allosteric inhibitor TH-470 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-methylpropanoylamino)-~{N}-[2-[(phenylmethyl)-[4-(phenylsulfamoyl)phenyl]carbonyl-amino]ethyl]-1,3-thiazole-5-carboxamide, LIM domain kinase 1 | | Authors: | Lee, H, Yosaatmadja, Y, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Elkins, J.M. | | Deposit date: | 2020-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of LIMK1 Kinase domain with allosteric inhibitor TH-470
To Be Published
|
|
7CW3
 
 | | Cryo-EM structure of Chikungunya virus in complex with mAb CHK-263 IgG (subregion around icosahedral 2-fold vertex) | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
