9EYS
 
 | | Structure of Far-Red Photosystem I from C. thermalis PCC 7203 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Consoli, G, Tufaill, F, Murray, J.W, Fantuzzi, A, Rutherford, A.W. | | Deposit date: | 2024-04-09 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.01 Å) | | Cite: | Structure of Far-Red Photosystem I from C. thermalis PCC 7203
To Be Published
|
|
9FDH
 
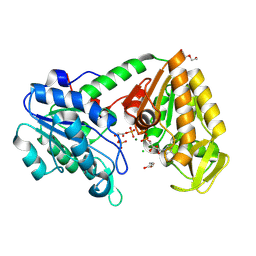 | | Closed Human phosphoglycerate kinase complex with BPG and ADP produced by cross-soaking a TSA crystal | | Descriptor: | 1,3-BISPHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Cliff, M.J, Waltho, J.P, Bowler, M.W, Baxter, N.J, Bisson, C, Blackburn, G.M. | | Deposit date: | 2024-05-17 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | The role of magnesium in catalysis by phosphoglycerate kinase
To be published
|
|
9FDN
 
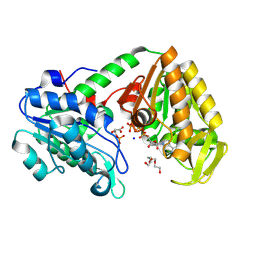 | | Human phosphoglycerate kinase in complex with ATP and 3PG formed by cross-soaking a TSA crystal | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Phosphoglycerate kinase 1, ... | | Authors: | Cliff, M.J, Waltho, J.P, Bowler, M.W, Baxter, N.J, Bisson, C, Blackburn, G.M. | | Deposit date: | 2024-05-17 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The role of magnesium in catalysis by phosphoglycerate kinase
To be published
|
|
9FFM
 
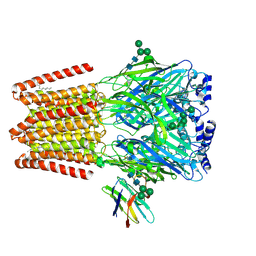 | |
9FF2
 
 | |
9FEZ
 
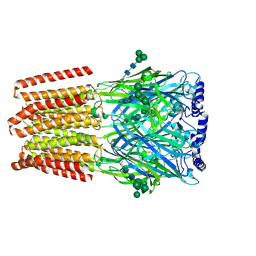 | |
9FFN
 
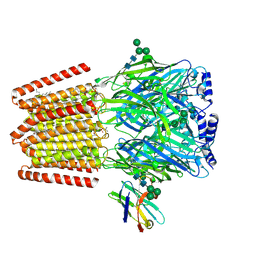 | |
9G3U
 
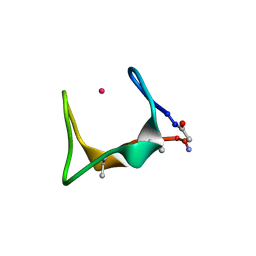 | |
9G3C
 
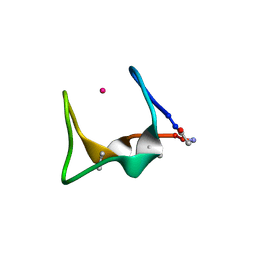 | |
9G39
 
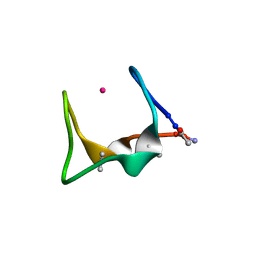 | |
9GBF
 
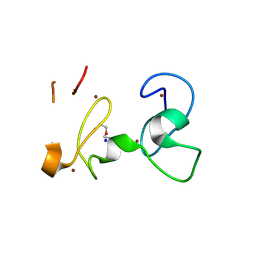 | | X-RAY structure of PHDvC5HCH tandem domain of NSD2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone-lysine N-methyltransferase NSD2, ... | | Authors: | Musco, G, Cocomazzi, P, Berardi, A, Knapp, S, Kramer, A. | | Deposit date: | 2024-07-31 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The C-terminal PHDVC5HCH tandem domain of NSD2 is a combinatorial reader of unmodified H3K4 and tri-methylated H3K27 that regulates transcription of cell adhesion genes in multiple myeloma.
Nucleic Acids Res., 53, 2025
|
|
9FUL
 
 | |
9FYU
 
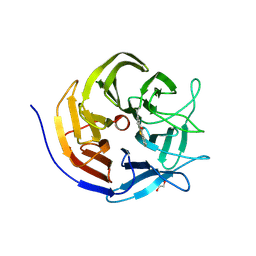 | |
9G8K
 
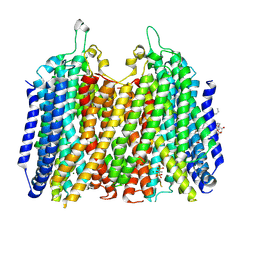 | | Structure of K+-dependent Na+-PPase from Thermotoga maritima in complex with Ca2+ and Etidronate | | Descriptor: | (1-hydroxyethane-1,1-diyl)bis(phosphonic acid), CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Vidilaseris, K, Liu, J, Goldman, A. | | Deposit date: | 2024-07-23 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Conformational dynamics and asymmetry in multimodal inhibition of membrane-bound pyrophosphatases
Elife, 2024
|
|
9GCA
 
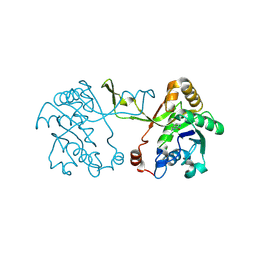 | |
9GFY
 
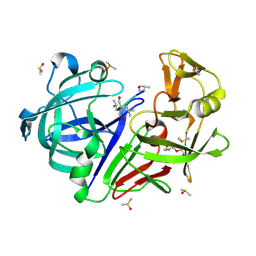 | | Endothiapepsin in complex with pepstatin soaked at pH 7.6 | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, PENTAETHYLENE GLYCOL, ... | | Authors: | Mueller, J.M, Glinca, S. | | Deposit date: | 2024-08-12 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Protonation Effects in Protein-Ligand Complexes - A Case Study of Endothiapepsin and Pepstatin A with Computational and Experimental Methods.
Chemmedchem, 20, 2025
|
|
9G6U
 
 | | p53-Y220C Core Domain Covalently Bound to 3,5-Dichloro-6-Ethylpyrazine-2-carbonitirle Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 3,5-bis(chloranyl)-6-ethyl-pyrazine-2-carbonitrile, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-19 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | SNAr Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes
Drug Des Devel Ther, 2025
|
|
9GC8
 
 | | Crystal structure of Pseudomonas aeruginosa IspD in complex with C11H12N2O3 | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, methyl 2-[(5~{R})-6-methyl-7-oxidanylidene-5~{H}-pyrrolo[3,4-b]pyridin-5-yl]ethanoate | | Authors: | Borel, F, D'Auria, L. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment Discovery by X-Ray Crystallographic Screening Targeting the CTP Binding Site of Pseudomonas Aeruginosa IspD.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9G6T
 
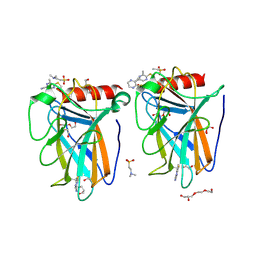 | | p53-Y220C Core Domain Covalently Bound to 5-Chloro-6-methylpyrazine-2-carbonitrile Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-chloranyl-6-methyl-pyrazine-2-carbonitrile, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-19 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | SNAr Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes
Drug Des Devel Ther, 2025
|
|
9H4A
 
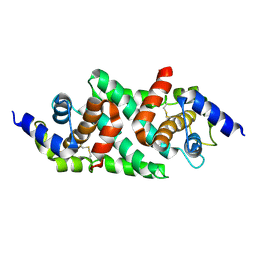 | | Ribonuclease-three Like 4 crystallization and structure determination at room temperature in the CrystalChip | | Descriptor: | Protein NUCLEAR FUSION DEFECTIVE 2 | | Authors: | Engilberge, S, Vincent, R, Coudray, L, Nilles, L, Scheer, H, Ritzenthaler, C, Sauter, C. | | Deposit date: | 2024-10-18 | | Release date: | 2024-12-11 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystallization and structure determination at room temperature in the CrystalChip.
Febs Open Bio, 15, 2025
|
|
9HKW
 
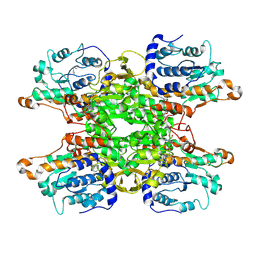 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 3 open and 1 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 3 open and 1 closed subunits
To Be Published
|
|
9HKY
 
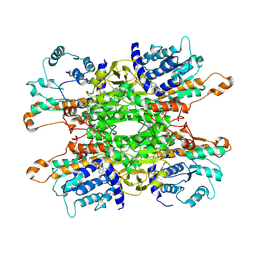 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 open and 2 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM struture of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 open and 2 closed subunits
To Be Published
|
|
9HKZ
 
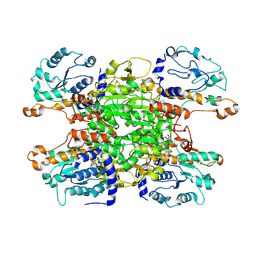 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 wide open, 1 open and 1 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 wide open, 1 open and 1 closed subunits
To Be Published
|
|
9HKX
 
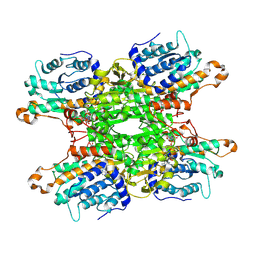 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 1 open and 3 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 1 open and 3 closed subunits
To Be Published
|
|
9HKV
 
 | |
