4LLJ
 
 | | Crystal structure of PDE10A2 with fragment ZT214 | | Descriptor: | 2H-isoindole-1,3-diamine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM2
 
 | | Crystal structure of PDE10A2 with fragment ZT462 | | Descriptor: | 2,3-dihydro-1,4-benzodioxin-6-ylmethanol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM0
 
 | | Crystal structure of PDE10A2 with fragment ZT448 | | Descriptor: | 5-NITROINDAZOLE, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LLK
 
 | | Crystal structure of PDE10A2 with fragment ZT217 | | Descriptor: | 2-methylquinazolin-4(3H)-one, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
3TVX
 
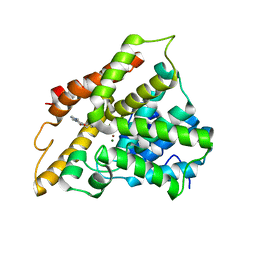 | | The structure of PDE4A with pentoxifylline at 2.84A resolution | | Descriptor: | 3,7-DIMETHYL-1-(5-OXOHEXYL)-3,7-DIHYDRO-1H-PURINE-2,6-DIONE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Badger, J, Sridhar, V. | | Deposit date: | 2011-09-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Fragment-Based Screening for Inhibitors of PDE4A Using Enthalpy Arrays and X-ray Crystallography.
J Biomol Screen, 17, 2012
|
|
3SZ2
 
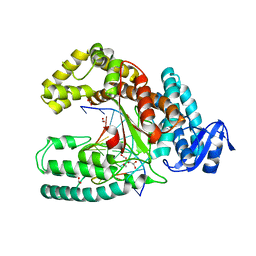 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dG as templating nucleobase | | Descriptor: | (5'-D(*AP*AP*AP*GP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3'), DNA polymerase I, ... | | Authors: | Betz, K, Marx, A, Diederichs, K. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3SIS
 
 | |
3LM5
 
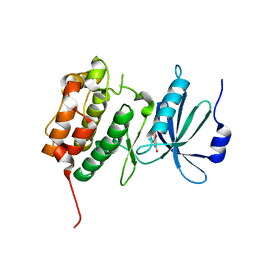 | | Crystal Structure of human Serine/Threonine Kinase 17B (STK17B) in complex with Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Serine/threonine-protein kinase 17B | | Authors: | Ugochukwu, E, Soundararajan, M, Rellos, P, Fedorov, O, Phillips, C, Wang, J, Hapka, E, Filippakopoulos, P, Chaikuad, A, Pike, A.C.W, Carpenter, L, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
3UW6
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR120 | | Descriptor: | Alanine racemase | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-11-30 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of enone-binding proteins with catalytic activity for the Morita-Baylis-Hillman reaction.
Acs Chem.Biol., 8, 2013
|
|
3SV4
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dT as templating nucleobase | | Descriptor: | (5'-D(*AP*AP*AP*TP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA polymerase I, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3SYZ
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dNaM as templating nucleobase | | Descriptor: | (5'-D(*AP*AP*AP*(BMN)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA polymerase I, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3TB0
 
 | |
3TAY
 
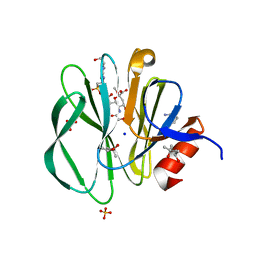 | |
3SV3
 
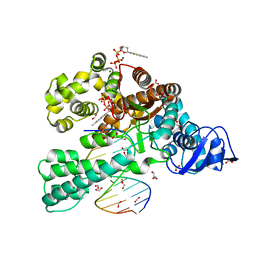 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with the artificial base pair dNaM-d5SICSTP | | Descriptor: | (5'-D(*AP*AP*AP*(BMN)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), 2-{2-deoxy-5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-6-methylisoquinoline-1(2H)-thione, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
3U26
 
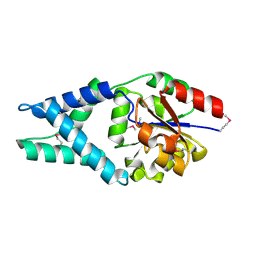 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR48 | | Descriptor: | PF00702 domain protein | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Computational design of enone-binding proteins with catalytic activity for the Morita-Baylis-Hillman reaction.
Acs Chem.Biol., 8, 2013
|
|
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | Descriptor: | Transforming growth factor beta-2 proprotein | | Authors: | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
1SF0
 
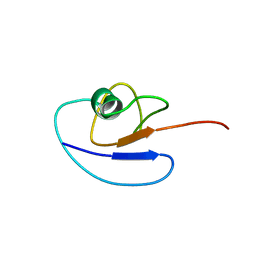 | |
1RWS
 
 | |
2P3I
 
 | | Crystal structure of Rhesus Rotavirus VP8* at 295K | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, SULFATE ION, VP4 | | Authors: | Blanchard, H. | | Deposit date: | 2007-03-09 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein
Glycobiology, 19, 2009
|
|
2P3K
 
 | | Crystal structure of Rhesus rotavirus VP8* at 100K | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Blanchard, H. | | Deposit date: | 2007-03-09 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein
Glycobiology, 19, 2009
|
|
2P3J
 
 | |
2QVT
 
 | | Structure of Melampsora lini avirulence protein, AvrL567-D | | Descriptor: | AvrL567-D | | Authors: | Guncar, G, Wang, C.I, Forwood, J.K, Teh, T, Catanzariti, A.M, Lawrence, G, Schirra, H.J, Anderson, P.A, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity.
Plant Cell, 19, 2007
|
|
3ZRH
 
 | | Crystal structure of the Lys29, Lys33-linkage-specific TRABID OTU deubiquitinase domain reveals an Ankyrin-repeat ubiquitin binding domain (AnkUBD) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UBIQUITIN THIOESTERASE ZRANB1 | | Authors: | Licchesi, J.D.F, Akutsu, M, Komander, D. | | Deposit date: | 2011-06-16 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | An Ankyrin-Repeat Ubiquitin-Binding Domain Determines Trabid'S Specificity for Atypical Ubiquitin Chains.
Nat.Struct.Mol.Biol., 19, 2011
|
|
4XYJ
 
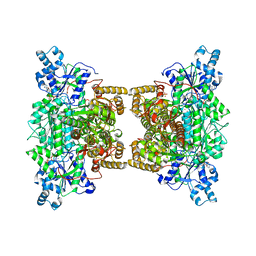 | | Crystal structure of human phosphofructokinase-1 in complex with ATP and Mg, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
3I1C
 
 | |
