1C8G
 
 | | FELINE PANLEUKOPENIA VIRUS EMPTY CAPSID STRUCTURE | | Descriptor: | CALCIUM ION, FELINE PANLEUKOPENIA VIRUS CAPSID | | Authors: | Rossmann, M.G, Simpson, A.A. | | Deposit date: | 2000-05-05 | | Release date: | 2000-08-09 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses.
J.Mol.Biol., 300, 2000
|
|
1C8D
 
 | |
1C8H
 
 | |
1IJS
 
 | | CPV (STRAIN D) mutant A300D, complex (VIRAL COAT/DNA), VP2, PH=7.5, T=4 DEGREES C | | Descriptor: | DNA (5'-D(*AP*C)-3'), DNA (5'-D(*CP*CP*AP*CP*CP*CP*CP*AP*A)-3'), PROTEIN (PARVOVIRUS COAT PROTEIN) | | Authors: | Llamas-Saiz, A.L, Agbandje-McKenna, M, Parker, J.S.L, Wahid, A.T.M, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 1996-09-12 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range.
Virology, 225, 1996
|
|
7QZD
 
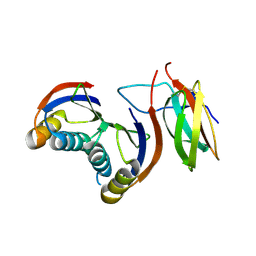 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikF with an engineered HMA domain of Pikp-1 (Pikp-SNK-EKE) from rice (Oryza sativa) | | Descriptor: | Avr-Pik, Resistance protein Pikp-1 | | Authors: | Maidment, J.H.R, Franceschetti, M, Longya, A, Banfield, M.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effector target-guided engineering of an integrated domain expands the disease resistance profile of a rice NLR immune receptor.
Elife, 12, 2023
|
|
1P5Y
 
 | |
1P5W
 
 | |
7QPX
 
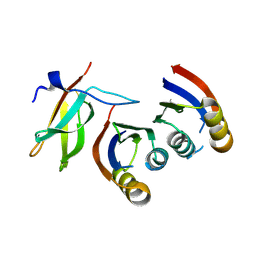 | |
1DNV
 
 | | PARVOVIRUS (DENSOVIRUS) FROM GALLERIA MELLONELLA | | Descriptor: | GALLERIA MELLONELLA DENSOVIRUS CAPSID PROTEIN | | Authors: | Simpson, A.A, Chipmann, P.R, Baker, T.S, Tijssen, P, Rossmann, M.G. | | Deposit date: | 1998-07-22 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 A resolution.
Structure, 6, 1998
|
|
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
7PC2
 
 | | HIV-1 Env (BG505 SOSIP.664) in complex with the IgA bNAb 7-269 and the antibody 3BNC117. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC IgG Fab heavy chain, ... | | Authors: | Fernandez, I, Bontems, F, Pehau-Arnaudet, G, Rey, F. | | Deposit date: | 2021-08-03 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Epitope convergence of broadly HIV-1 neutralizing IgA and IgG antibody lineages in a viremic controller.
J.Exp.Med., 219, 2022
|
|
6X9I
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
6X9K
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3685032A | | Descriptor: | (2R)-2-{[6-(4-aminopiperidin-1-yl)-3,5-dicyano-4-ethylpyridin-2-yl]sulfanyl}-2-phenylacetamide, 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
6X9J
 
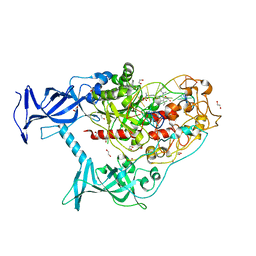 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3830052 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
4XTQ
 
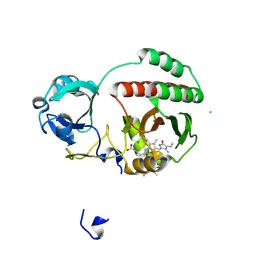 | | Crystal structure of a mutant (C20S) of a near-infrared fluorescent protein BphP1-FP | | Descriptor: | 3-[2-[(Z)-[5-[(Z)-[(3R,4R)-3-ethenyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, BphP1-FP/C20S, CHLORIDE ION | | Authors: | Pletnev, S, Malashkevich, V.N. | | Deposit date: | 2015-01-23 | | Release date: | 2015-12-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Molecular Basis of Spectral Diversity in Near-Infrared Phytochrome-Based Fluorescent Proteins.
Chem.Biol., 22, 2015
|
|
6FDG
 
 | |
6EZ6
 
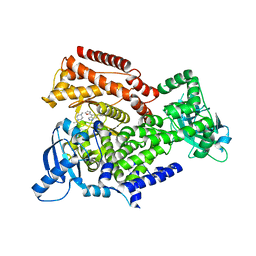 | | PI3 kinase delta in complex with Methyl 5-(4-(5-((4-isopropylpiperazin-1-yl)methyl)oxazol-2-yl)-1H-indazol-6-yl)-2-methoxynicotinate | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, methyl 2-methoxy-5-[4-[5-[(4-propan-2-ylpiperazin-1-yl)methyl]-1,3-oxazol-2-yl]-2~{H}-indazol-6-yl]pyridine-3-carboxylate | | Authors: | Convery, M.A, Campos, S, Dalton, S.E. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Selectively Targeting the Kinome-Conserved Lysine of PI3K delta as a General Approach to Covalent Kinase Inhibition.
J. Am. Chem. Soc., 140, 2018
|
|
6EYZ
 
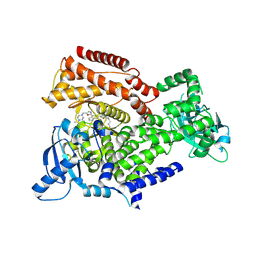 | | PI3 kinase delta in complex with 4-Fluorophenyl 5-(4-(5-((4-isopropylpiperazin-1-yl)methyl)oxazol-2-yl)-1H-indazol-6-yl)-2-methoxynicotinate | | Descriptor: | 2-methoxy-5-[4-[5-[(4-propan-2-ylpiperazin-1-yl)methyl]-1,3-oxazol-2-yl]-2~{H}-indazol-6-yl]pyridine-3-carboxylic acid, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Campos, S, Dalton, S.E. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-20 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectively Targeting the Kinome-Conserved Lysine of PI3K delta as a General Approach to Covalent Kinase Inhibition.
J. Am. Chem. Soc., 140, 2018
|
|
5MTO
 
 | | N-terminal domain of the human tumor suppressor ING5 C19S mutant | | Descriptor: | Inhibitor of growth protein 5, SODIUM ION, SULFATE ION | | Authors: | Ormaza, G, Buitrago, J.A.R, Roversi, P, Rojas, A.L, Blanco, F.J. | | Deposit date: | 2017-01-10 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Tumor Suppressor ING5 Is a Dimeric, Bivalent Recognition Molecule of the Histone H3K4me3 Mark.
J.Mol.Biol., 431, 2019
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
3KZZ
 
 | |
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
